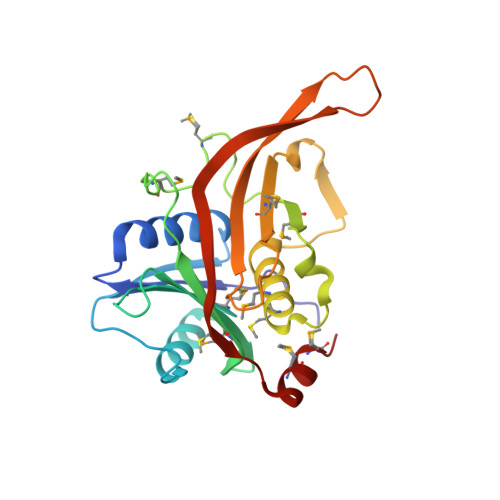
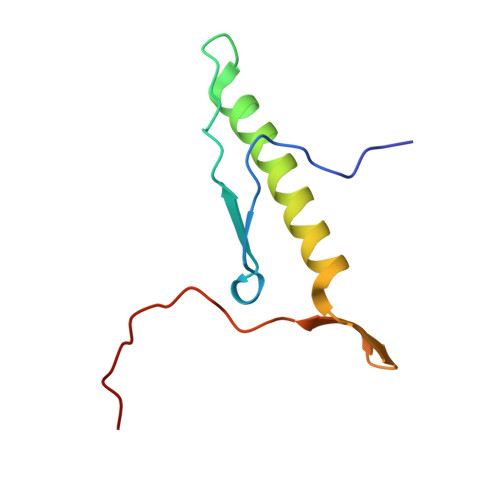
The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis.
Kharde, S., Calvino, F.R., Gumiero, A., Wild, K., Sinning, I.(2015) Nucleic Acids Res 43: 7083-7095
- PubMed: 26117542
- DOI: https://doi.org/10.1093/nar/gkv640
- Primary Citation of Related Structures:
5BY8 - PubMed Abstract:
The assembly of eukaryotic ribosomes is a hierarchical process involving about 200 biogenesis factors and a series of remodeling steps. The 5S RNP consisting of the 5S rRNA, RpL5 and RpL11 is recruited at an early stage, but has to rearrange during maturation of the pre-60S ribosomal subunit. Rpf2 and Rrs1 have been implicated in 5S RNP biogenesis, but their precise role was unclear. Here, we present the crystal structure of the Rpf2-Rrs1 complex from Aspergillus nidulans at 1.5 Å resolution and describe it as Brix domain of Rpf2 completed by Rrs1 to form two anticodon-binding domains with functionally important tails. Fitting the X-ray structure into the cryo-EM density of a previously described pre-60S particle correlates with biochemical data. The heterodimer forms specific contacts with the 5S rRNA, RpL5 and the biogenesis factor Rsa4. The flexible protein tails of Rpf2-Rrs1 localize to the central protuberance. Two helices in the Rrs1 C-terminal tail occupy a strategic position to block the rotation of 25S rRNA and the 5S RNP. Our data provide a structural model for 5S RNP recruitment to the pre-60S particle and explain why removal of Rpf2-Rrs1 is necessary for rearrangements to drive 60S maturation.
- Heidelberg University Biochemistry Center (BZH), INF 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: