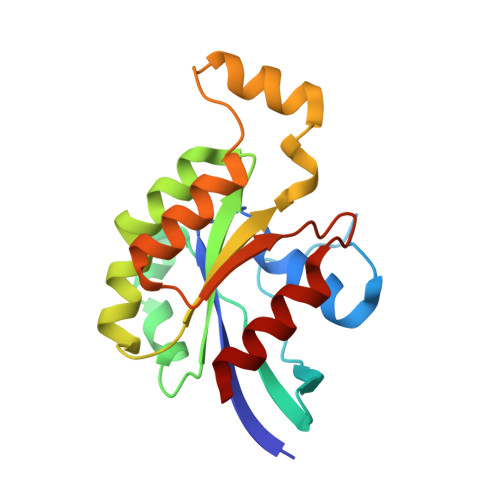
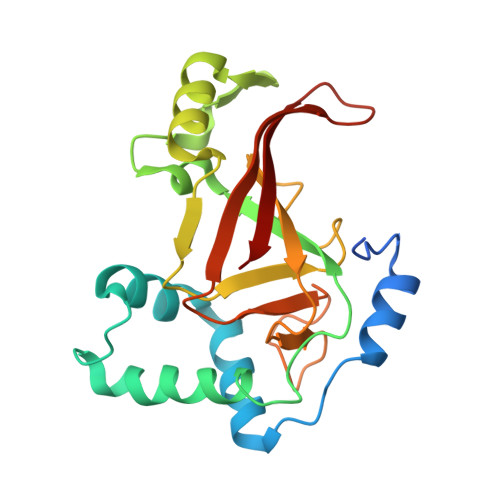
Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
Toda, A., Tsurumura, T., Yoshida, T., Tsumori, Y., Tsuge, H.(2015) J Biological Chem 290: 19423-19432
- PubMed: 26067270
- DOI: https://doi.org/10.1074/jbc.M115.653220
- Primary Citation of Related Structures:
4XSG, 4XSH, 5BWM - PubMed Abstract:
C3 exoenzyme is a mono-ADP-ribosyltransferase (ART) that catalyzes transfer of an ADP-ribose moiety from NAD(+) to Rho GTPases. C3 has long been used to study the diverse regulatory functions of Rho GTPases. How C3 recognizes its substrate and how ADP-ribosylation proceeds are still poorly understood. Crystal structures of C3-RhoA complex reveal that C3 recognizes RhoA via the switch I, switch II, and interswitch regions. In C3-RhoA(GTP) and C3-RhoA(GDP), switch I and II adopt the GDP and GTP conformations, respectively, which explains why C3 can ADP-ribosylate both nucleotide forms. Based on structural information, we successfully changed Cdc42 to an active substrate with combined mutations in the C3-Rho GTPase interface. Moreover, the structure reflects the close relationship among Gln-183 in the QXE motif (C3), a modified Asn-41 residue (RhoA) and NC1 of NAD(H), which suggests that C3 is the prototype ART. These structures show directly for the first time that the ARTT loop is the key to target protein recognition, and they also serve to bridge the gaps among independent studies of Rho GTPases and C3.
- From the Department of Bioresource and Environmental Sciences, Faculty of Life Sciences, and.
Organizational Affiliation: