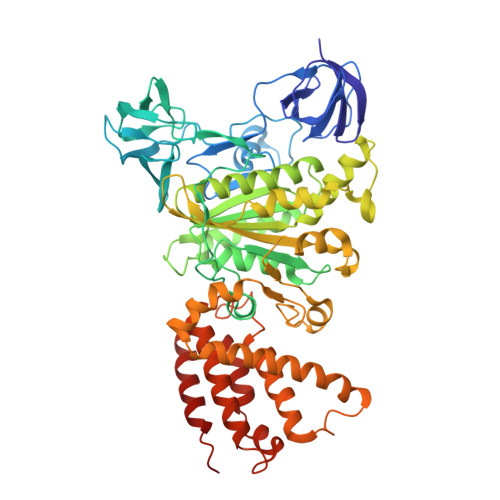
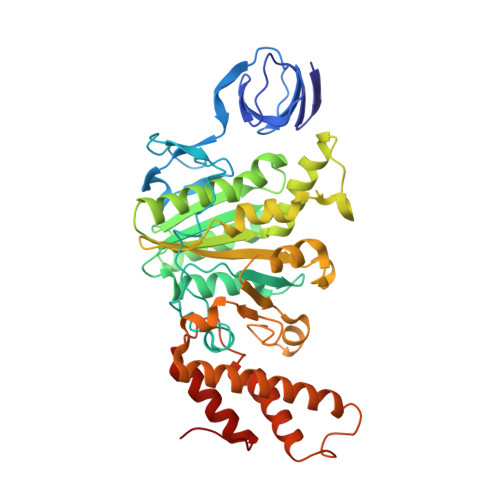
Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
Mohanty, S., Jobichen, C., Chichili, V.P.R., Velazquez-Campoy, A., Low, B.C., Hogue, C.W.V., Sivaraman, J.(2015) J Biological Chem 290: 27280-27296
- PubMed: 26370083
- DOI: https://doi.org/10.1074/jbc.M115.677492
- Primary Citation of Related Structures:
5BN3, 5BN4, 5BN5, 5BO5 - PubMed Abstract:
ATP synthesis is a critical and universal life process carried out by ATP synthases. Whereas eukaryotic and prokaryotic ATP synthases are well characterized, archaeal ATP synthases are relatively poorly understood. The hyperthermophilic archaeal parasite, Nanoarcheaum equitans, lacks several subunits of the ATP synthase and is suspected to be energetically dependent on its host, Ignicoccus hospitalis. This suggests that this ATP synthase might be a rudimentary machine. Here, we report the crystal structures and biophysical studies of the regulatory subunit, NeqB, the apo-NeqAB, and NeqAB in complex with nucleotides, ADP, and adenylyl-imidodiphosphate (non-hydrolysable analog of ATP). NeqB is ∼20 amino acids shorter at its C terminus than its homologs, but this does not impede its binding with NeqA to form the complex. The heterodimeric NeqAB complex assumes a closed, rigid conformation irrespective of nucleotide binding; this differs from its homologs, which require conformational changes for catalytic activity. Thus, although N. equitans possesses an ATP synthase core A3B3 hexameric complex, it might not function as a bona fide ATP synthase.
- Department of Biological Sciences, National University of Singapore, Singapore 117543, Singapore.
Organizational Affiliation: