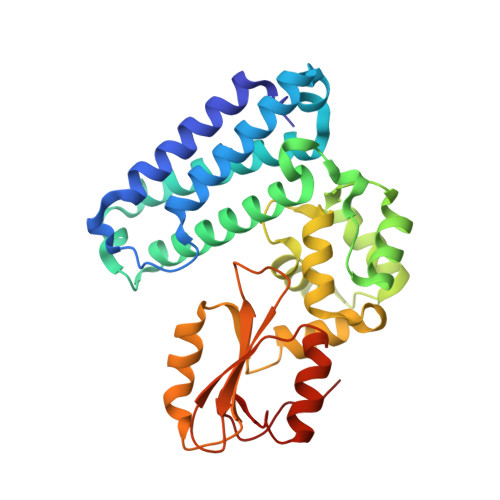
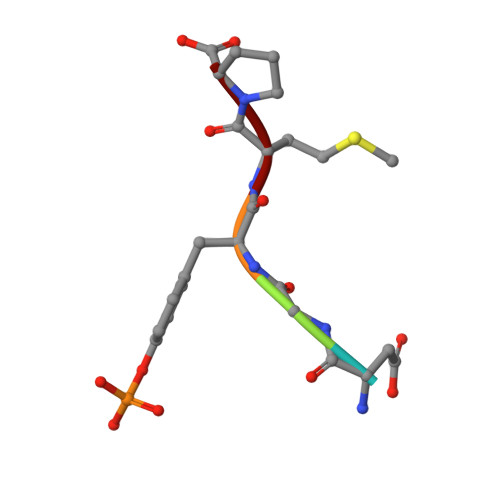
Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Ohno, A., Ochi, A., Maita, N., Ueji, T., Bando, A., Nakao, R., Hirasaka, K., Abe, T., Teshima-Kondo, S., Nemoto, H., Okumura, Y., Higashibata, A., Yano, S., Tochio, H., Nikawa, T.(2016) Arch Biochem Biophys 594: 1-7
- PubMed: 26874193
- DOI: https://doi.org/10.1016/j.abb.2016.02.014
- Primary Citation of Related Structures:
5AXI - PubMed Abstract:
Cbl-b is a RING-type ubiquitin ligase. Previously, we showed that Cbl-b-mediated ubiquitination and proteosomal degradation of IRS-1 contribute to muscle atrophy caused by unloading stress. The phospho-pentapeptide DGpYMP (Cblin) mimics Tyr612-phosphorylated IRS-1 and inhibits the Cbl-b-mediated ubiquitination and degradation of IRS-1 in vitro and in vivo. In this study, we confirmed the direct interaction between Cblin and the TKB domain of Cbl-b using NMR. Moreover, we showed that the shortened tripeptide GpYM also binds to the TKB domain. To elucidate the inhibitory mechanism of Cblin, we solved the crystal structure of the TKB-Cblin complex at a resolution of 2.5 Å. The pY in Cblin inserts into a positively charged pocket in the TKB domain via hydrogen-bond networks and hydrophobic interactions. Within this complex, the Cblin structure closely resembles the TKB-bound form of another substrate-derived phosphopeptide, Zap-70-derived phosphopeptide. These peptides lack the conserved intrapeptidyl hydrogen bond between pY and a conserved residue involved in TKB-domain binding. Instead of the conserved interaction, these peptides specifically interact with the TKB domain. Based on this binding mode of Cblin to the TKB domain, we can design drugs against unloading-mediated muscle atrophy.
- Departments of Nutritional Physiology, Institute of Health Biosciences, University of Tokushima Graduate School, Tokushima, 770-8503, Japan.
Organizational Affiliation: