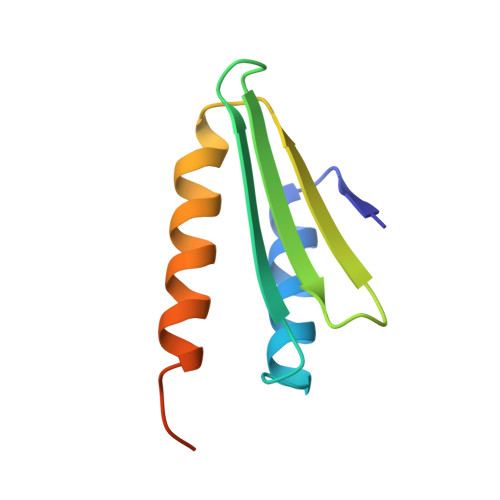
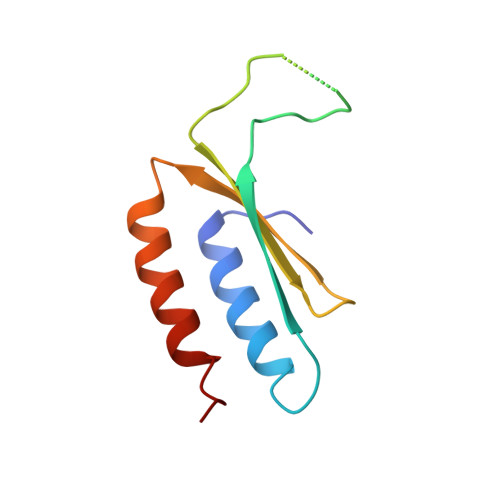
Retrotransposition and Crystal Structure of an Alu Rnp in the Ribosome-Stalling Conformation.
Ahl, V., Keller, H., Schmidt, S., Weichenrieder, O.(2015) Mol Cell 60: 715
- PubMed: 26585389
- DOI: https://doi.org/10.1016/j.molcel.2015.10.003
- Primary Citation of Related Structures:
5AOX - PubMed Abstract:
The Alu element is the most successful human genomic parasite affecting development and causing disease. It originated as a retrotransposon during early primate evolution of the gene encoding the signal recognition particle (SRP) RNA. We defined a minimal Alu RNA sufficient for effective retrotransposition and determined a high-resolution structure of its complex with the SRP9/14 proteins. The RNA adopts a compact, closed conformation that matches the envelope of the SRP Alu domain in the ribosomal translation elongation factor-binding site. Conserved structural elements in SRP RNAs support an ancient function of the closed conformation that predates SRP9/14. Structure-based mutagenesis shows that retrotransposition requires the closed conformation of the Alu ribonucleoprotein particle and is consistent with the recognition of stalled ribosomes. We propose that ribosome stalling is a common cause for the cis-preference of the mammalian L1 retrotransposon and for the efficiency of the Alu RNA in hijacking nascent L1 reverse transcriptase.
- Department of Biochemistry, Max Planck Institute for Developmental Biology, Spemannstrasse 35, 72076 Tübingen, Germany.
Organizational Affiliation: