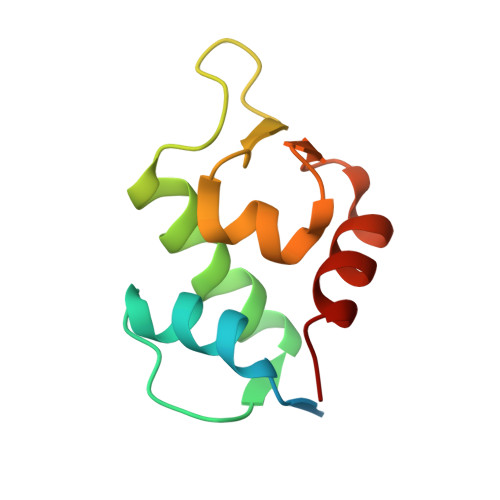
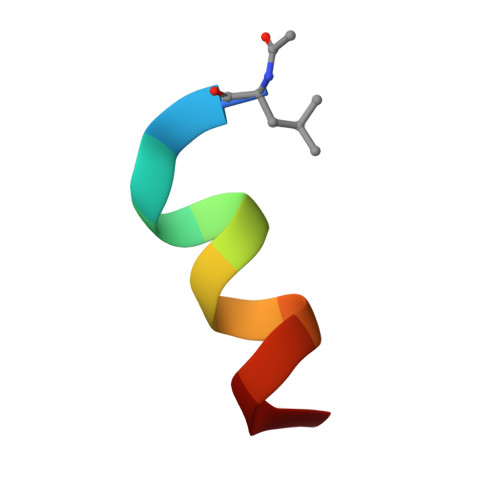
Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Lau, Y.H., Wu, Y., Rossmann, M., Tan, B.X., De Andrade, P., Tan, Y.S., Verma, C., Mckenzie, G.J., Venkitaraman, A.R., Hyvonen, M., Spring, D.R.(2015) Angew Chem Int Ed Engl 54: 15410
- PubMed: 26768531
- DOI: https://doi.org/10.1002/anie.201508416
- Primary Citation of Related Structures:
5AFG - PubMed Abstract:
Peptide stapling is a method for designing macrocyclic alpha-helical inhibitors of protein-protein interactions. However, obtaining a cell-active inhibitor can require significant optimization. We report a novel stapling technique based on a double strain-promoted azide-alkyne reaction, and exploit its biocompatibility to accelerate the discovery of cell-active stapled peptides. As a proof of concept, MDM2-binding peptides were stapled in parallel, directly in cell culture medium in 96-well plates, and simultaneously evaluated in a p53 reporter assay. This in situ stapling/screening process gave an optimal candidate that showed improved proteolytic stability and nanomolar binding to MDM2 in subsequent biophysical assays. α-Helicity was confirmed by a crystal structure of the MDM2-peptide complex. This work introduces in situ stapling as a versatile biocompatible technique with many other potential high-throughput biological applications.
- Department of Chemistry, University of Cambridge, Lensfield Rd, Cambridge CB2 1EW (UK).
Organizational Affiliation: