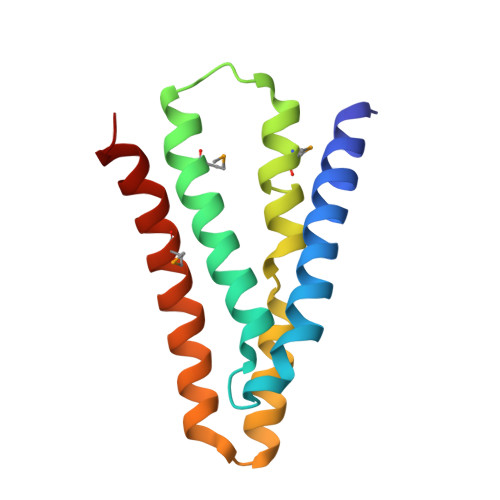
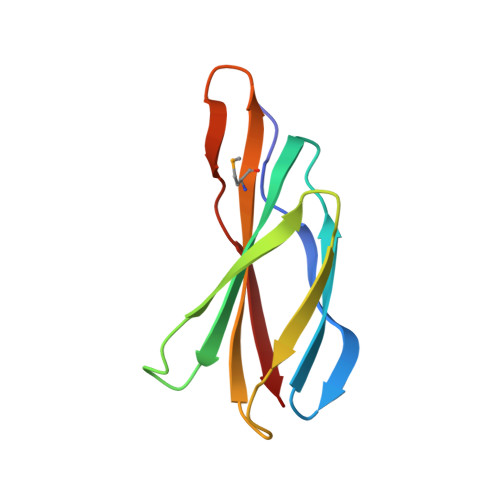
Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Stockbridge, R.B., Kolmakova-Partensky, L., Shane, T., Koide, A., Koide, S., Miller, C., Newstead, S.(2015) Nature 525: 548
- PubMed: 26344196
- DOI: https://doi.org/10.1038/nature14981
- Primary Citation of Related Structures:
5A40, 5A43, 5NKQ - PubMed Abstract:
To contend with hazards posed by environmental fluoride, microorganisms export this anion through F(-)-specific ion channels of the Fluc family. Since the recent discovery of Fluc channels, numerous idiosyncratic features of these proteins have been unearthed, including strong selectivity for F(-) over Cl(-) and dual-topology dimeric assembly. To understand the chemical basis for F(-) permeation and how the antiparallel subunits convene to form a F(-)-selective pore, here we solve the crystal structures of two bacterial Fluc homologues in complex with three different monobody inhibitors, with and without F(-) present, to a maximum resolution of 2.1 Å. The structures reveal a surprising 'double-barrelled' channel architecture in which two F(-) ion pathways span the membrane, and the dual-topology arrangement includes a centrally coordinated cation, most likely Na(+). F(-) selectivity is proposed to arise from the very narrow pores and an unusual anion coordination that exploits the quadrupolar edges of conserved phenylalanine rings.
- Department of Biochemistry, Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts 02454, USA.
Organizational Affiliation: