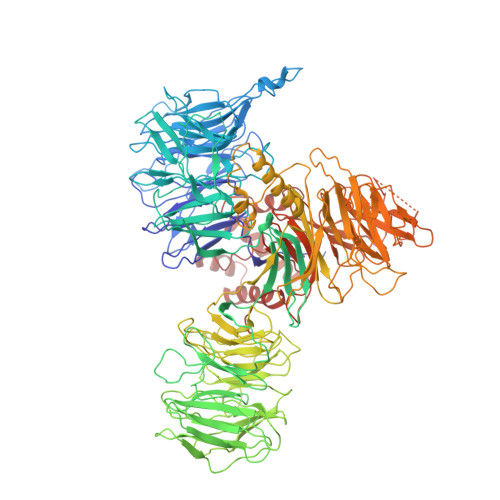
The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Finci, L.I., Zhang, X., Huang, X., Zhou, Q., Tsai, J., Teng, T., Agrawal, A., Chan, B., Irwin, S., Karr, C., Cook, A., Zhu, P., Reynolds, D., Smith, P.G., Fekkes, P., Buonamici, S., Larsen, N.A.(2018) Genes Dev 32: 309-320
- PubMed: 29491137
- DOI: https://doi.org/10.1101/gad.311043.117
- Primary Citation of Related Structures:
5ZYA - PubMed Abstract:
Somatic mutations in spliceosome proteins lead to dysregulated RNA splicing and are observed in a variety of cancers. These genetic aberrations may offer a potential intervention point for targeted therapeutics. SF3B1, part of the U2 small nuclear RNP (snRNP), is targeted by splicing modulators, including E7107, the first to enter clinical trials, and, more recently, H3B-8800. Modulating splicing represents a first-in-class opportunity in drug discovery, and elucidating the structural basis for the mode of action opens up new possibilities for structure-based drug design. Here, we present the cryogenic electron microscopy (cryo-EM) structure of the SF3b subcomplex (SF3B1, SF3B3, PHF5A, and SF3B5) bound to E7107 at 3.95 Å. This structure shows that E7107 binds in the branch point adenosine-binding pocket, forming close contacts with key residues that confer resistance upon mutation: SF3B1 R1074H and PHF5A Y36C The structure suggests a model in which splicing modulators interfere with branch point adenosine recognition and supports a substrate competitive mechanism of action (MOA). Using several related chemical probes, we validate the pose of the compound and support their substrate competitive MOA by comparing their activity against both strong and weak pre-mRNA substrates. Finally, we present functional data and structure-activity relationship (SAR) on the PHF5A R38C mutation that sensitizes cells to some chemical probes but not others. Developing small molecule splicing modulators represents a promising therapeutic approach for a variety of diseases, and this work provides a significant step in enabling structure-based drug design for these elaborate natural products. Importantly, this work also demonstrates that the utilization of cryo-EM in drug discovery is coming of age.
- Beijing Advanced Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: