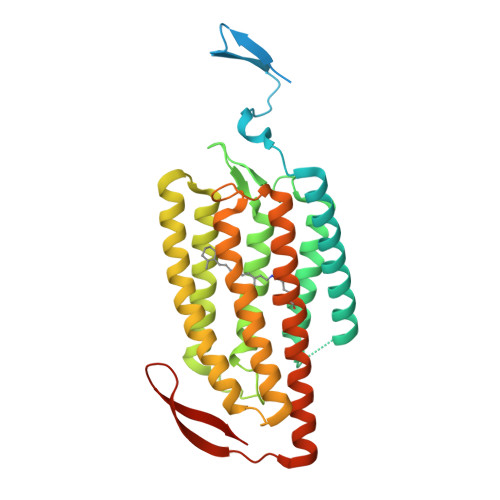
Crystal structure of the red light-activated channelrhodopsin Chrimson.
Oda, K., Vierock, J., Oishi, S., Rodriguez-Rozada, S., Taniguchi, R., Yamashita, K., Wiegert, J.S., Nishizawa, T., Hegemann, P., Nureki, O.(2018) Nat Commun 9: 3949-3949
- PubMed: 30258177
- DOI: https://doi.org/10.1038/s41467-018-06421-9
- Primary Citation of Related Structures:
5ZIH - PubMed Abstract:
Channelrhodopsins are light-activated ion channels that mediate cation permeation across cell membranes upon light absorption. Red-light-activated channelrhodopsins are of particular interest, because red light penetrates deeper into biological tissues and also enables dual-color experiments in combination with blue-light-activated optogenetic tools. Here we report the crystal structure of the most red-shifted channelrhodopsin from the algae Chlamydomonas noctigama, Chrimson, at 2.6 Å resolution. Chrimson resembles prokaryotic proton pumps in the retinal binding pocket, while sharing similarity with other channelrhodopsins in the ion-conducting pore. Concomitant mutation analysis identified the structural features that are responsible for Chrimson's red light sensitivity; namely, the protonation of the counterion for the retinal Schiff base, and the polar residue distribution and rigidity of the retinal binding pocket. Based on these mechanistic insights, we engineered ChrimsonSA, a mutant with a maximum activation wavelength red-shifted beyond 605 nm and accelerated closing kinetics.
- Department of Biological Sciences Graduate School of Science, The University of Tokyo, Tokyo, 113-0034, Japan.
Organizational Affiliation: