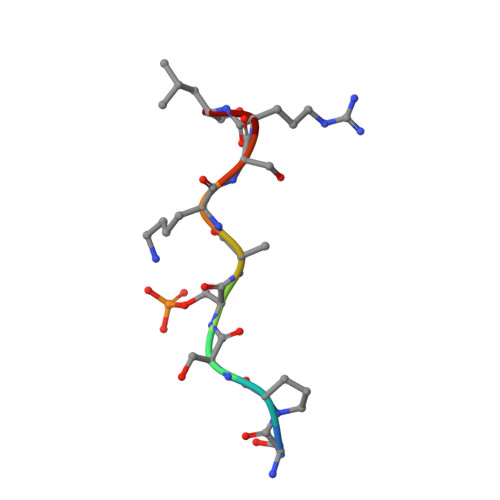
Structural Basis for Recognition of a Unique Epitope by a Human Anti-tau Antibody.
Zhang, H., Zhu, X., Pascual, G., Wadia, J.S., Keogh, E., Hoozemans, J.J., Siregar, B., Inganas, H., Stoop, E.J.M., Goudsmit, J., Apetri, A., Koudstaal, W., Wilson, I.A.(2018) Structure 26: 1626-1634.e4
- PubMed: 30318466
- DOI: https://doi.org/10.1016/j.str.2018.08.012
- Primary Citation of Related Structures:
5ZIA - PubMed Abstract:
Aggregation of the hyperphosphorylated protein tau into neurofibrillary tangles and neuropil threads is a hallmark of Alzheimer disease (AD). Identification and characterization of the epitopes recognized by anti-tau antibodies might shed light on the molecular mechanisms of AD pathogenesis. Here we report on the biochemical and structural characterization of a tau-specific monoclonal antibody CBTAU-24.1, which was isolated from the human memory B cell repertoire. Immunohistochemical staining with CBTAU-24.1 specifically detects pathological tau structures in AD brain samples. The crystal structure of CBTAU-24.1 Fab with a phosphorylated tau peptide revealed recognition of a unique epitope (Ser235-Leu243) in the tau proline-rich domain. Interestingly, the antibody can bind tau regardless of phosphorylation state of its epitope region and also recognizes both monomeric and paired helical filament tau irrespective of phosphorylation status. This human anti-tau antibody and its unique epitope may aid in development of diagnostics and/or therapeutic AD strategies.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA; Beijing Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences, Beijing 100049, People's Republic of China.
Organizational Affiliation: