Structural characterization of a novel KH-domain containing plant chloroplast endonuclease
Rout, A.K., Singh, H., Patel, S., Raghvan, V., Gautam, S., Minda, R., Rao, B.J., Chary, K.V.R.(2018) Sci Rep 8: 13750-13750
- PubMed: 30214061
- DOI: https://doi.org/10.1038/s41598-018-31142-w
- Primary Citation of Related Structures:
5ZB6 - PubMed Abstract:
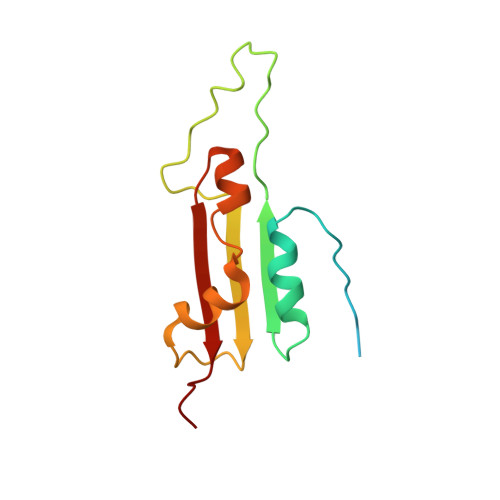
Chlamydomonas reinhardtii is a single celled alga that undergoes apoptosis in response to UV-C irradiation. UVI31+, a novel UV-inducible DNA endonuclease in C. reinhardtii, which normally localizes near cell wall and pyrenoid regions, gets redistributed into punctate foci within the whole chloroplast, away from the pyrenoid, upon UV-stress. Solution NMR structure of the first putative UV inducible endonuclease UVI31+ revealed an α 1 -β 1 -β 2 -α 2 -α 3 -β 3 fold similar to BolA and type II KH-domain ubiquitous protein families. Three α-helices of UVI31+ constitute one side of the protein surface, which are packed to the other side, made of three-stranded β-sheet, with intervening hydrophobic residues. A twenty-three residues long polypeptide stretch (D54-H76) connecting β 1 and β 2 strands is found to be highly flexible. Interestingly, UVI31+ recognizes the DNA primarily through its β-sheet. We propose that the catalytic triad residues involving Ser114, His95 and Thr116 facilitate DNA endonuclease activity of UVI31+. Further, decreased endonuclease activity of the S114A mutant is consistent with the direct participation of Ser114 in the catalysis. This study provides the first structural description of a plant chloroplast endonuclease that is regulated by UV-stress response.
- Department of Chemical Sciences, Tata Institute of Fundamental Research, Mumbai, 400005, India.
Organizational Affiliation: