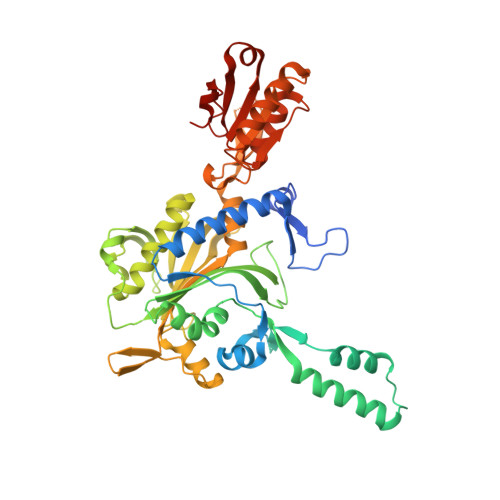
Glycyl-tRNA synthetase from Nanoarchaeum equitans: The first crystal structure of archaeal GlyRS and analysis of its tRNA glycylation.
Fujisawa, A., Toki, R., Miyake, H., Shoji, T., Doi, H., Hayashi, H., Hanabusa, R., Mutsuro-Aoki, H., Umehara, T., Ando, T., Noguchi, H., Voet, A., Park, S.Y., Tamura, K.(2019) Biochem Biophys Res Commun 511: 228-233
- PubMed: 30771900
- DOI: https://doi.org/10.1016/j.bbrc.2019.01.142
- Primary Citation of Related Structures:
5Z5E - PubMed Abstract:
This study reports the X-ray crystallographic structure of the glycyl-tRNA synthetase (GlyRS) of Nanoarchaeum equitans - a hyperthermophilic archaeal species. This is the first archaeal GlyRS crystal structure elucidated. The GlyRS comprises an N-terminal catalytic domain and a C-terminal anticodon-binding domain with a long β-sheet inserted between these domains. An unmodified transcript of the wild-type N. equitans tRNA Gly was successfully glycylated using GlyRS. Substitution of the discriminator base A73 of tRNA Gly with any other nucleotide caused a significant decrease in glycylation activity. Mutational analysis of the second base-pair C2G71 of the acceptor stem of tRNA Gly elucidated the importance of the base-pair, especially G71, as an identity element for recognition by GlyRS. Glycylation assays using tRNA Gly G71 substitution mutants and a GlyRS mutant where Arg223 is mutated to alanine strengthen the possibility that the carbonyl oxygen at position 6 of G71 would hydrogen-bond with the guanidine nitrogen of Arg223 in N. equitans GlyRS.
- Department of Biological Science and Technology, Tokyo University of Science, 6-3-1 Niijuku, Katsushika-ku, Tokyo 125-8585, Japan.
Organizational Affiliation: