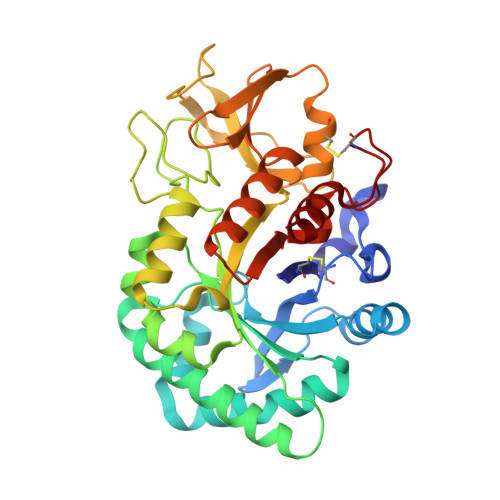
Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40
Sharma, P., Singh, P.K., Singh, N., Sharma, S., Kaur, P., Betzel, C., Singh, T.P.To be published.