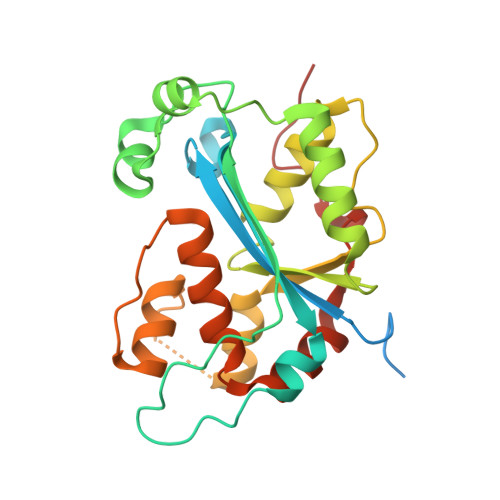
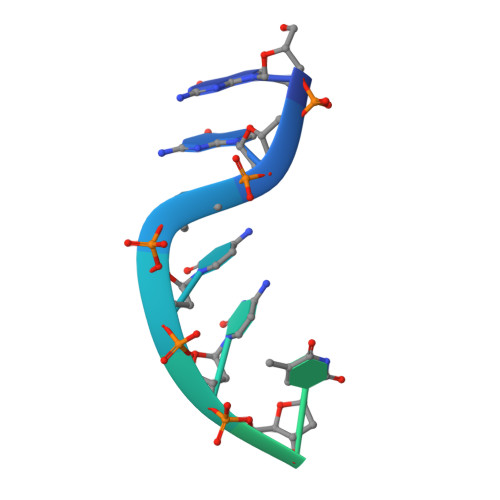
Structural basis for overhang excision and terminal unwinding of DNA duplexes by TREX1
Huang, K.W., Liu, T.C., Liang, R.Y., Chu, L.Y., Cheng, H.L., Chu, J.W., Hsiao, Y.Y.(2018) PLoS Biol 16: e2005653-e2005653
- PubMed: 29734329
- DOI: https://doi.org/10.1371/journal.pbio.2005653
- Primary Citation of Related Structures:
5YWS, 5YWT, 5YWU, 5YWV - PubMed Abstract:
Three prime repair exonuclease 1 (TREX1) is an essential exonuclease in mammalian cells, and numerous in vivo and in vitro data evidenced its participation in immunity regulation and in genotoxicity remediation. In these very complicated cellular functions, the molecular mechanisms by which duplex DNA substrates are processed are mostly elusive because of the lack of structure information. Here, we report multiple crystal structures of TREX1 complexed with various substrates to provide the structure basis for overhang excision and terminal unwinding of DNA duplexes. The substrates were designed to mimic the intermediate structural DNAs involved in various repair pathways. The results showed that the Leu24-Pro25-Ser26 cluster of TREX1 served to cap the nonscissile 5'-end of the DNA for precise removal of the short 3'-overhang in L- and Y-structural DNA or to wedge into the double-stranded region for further digestion along the duplex. Biochemical assays were also conducted to demonstrate that TREX1 can indeed degrade double-stranded DNA (dsDNA) to a full extent. Overall, this study provided unprecedented knowledge at the molecular level on the enzymatic substrate processing involved in prevention of immune activation and in responses to genotoxic stresses. For example, Arg128, whose mutation in TREX1 was linked to a disease state, were shown to exhibit consistent interaction patterns with the nonscissile strand in all of the structures we solved. Such structure basis is expected to play an indispensable role in elucidating the functional activities of TREX1 at the cellular level and in vivo.
- Department of Biological Science and Technology, National Chiao Tung University, Hsinchu, Taiwan, ROC.
Organizational Affiliation: