Crystal structures of unlinked full length NS3 from Dengue virus provide insights into dynamics of protease domain
Phoo, W.W., El Sahili, A., Zhang, Z.Z., Chen, M.W., Lescar, J., Vasudevan, S., Luo, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Genome polyprotein | A [auth B] | 599 | dengue virus type 4 | Mutation(s): 1 | 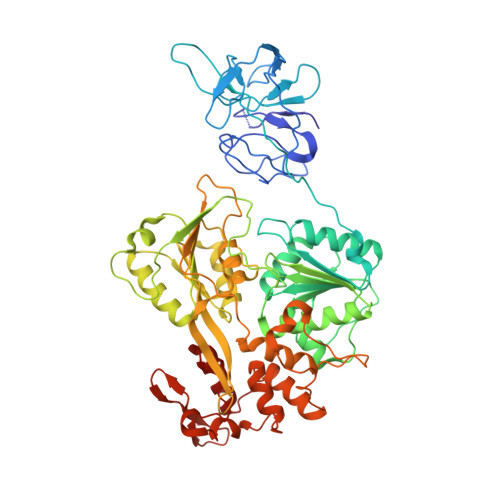 |
UniProt | |||||
Find proteins for Q2YHF0 (Dengue virus type 4 (strain Thailand/0348/1991)) Explore Q2YHF0 Go to UniProtKB: Q2YHF0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2YHF0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Genome polyprotein | B [auth A] | 47 | dengue virus type 4 | Mutation(s): 0 | 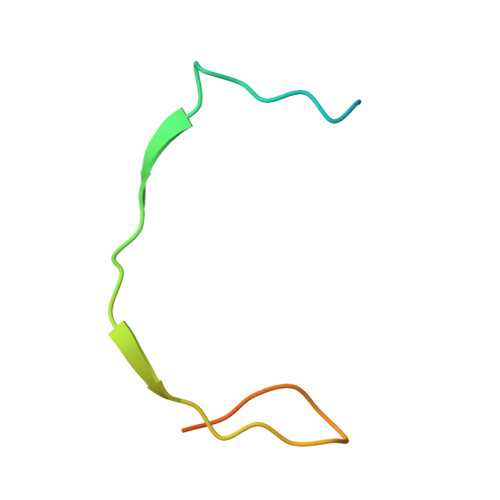 |
UniProt | |||||
Find proteins for Q2YHF0 (Dengue virus type 4 (strain Thailand/0348/1991)) Explore Q2YHF0 Go to UniProtKB: Q2YHF0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2YHF0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | C [auth B] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 52.77 | α = 90 |
| b = 88.3 | β = 92.34 |
| c = 81.11 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| NTU | Singapore | START UP GRANT |
| NMRC | Singapore | CBRG14MAY051 |
| NMRC | Singapore | CBRG/0103/2016 |
| NRF | Singapore | CRP001-063 |