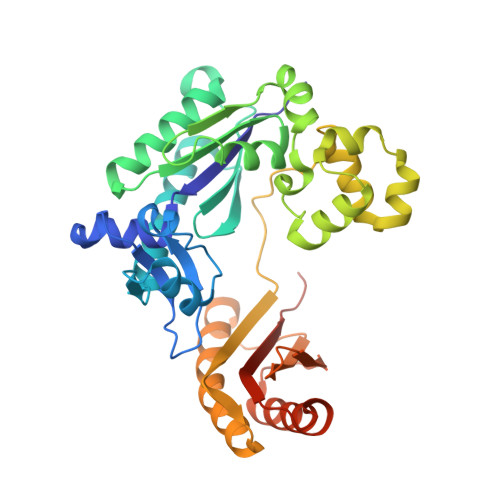
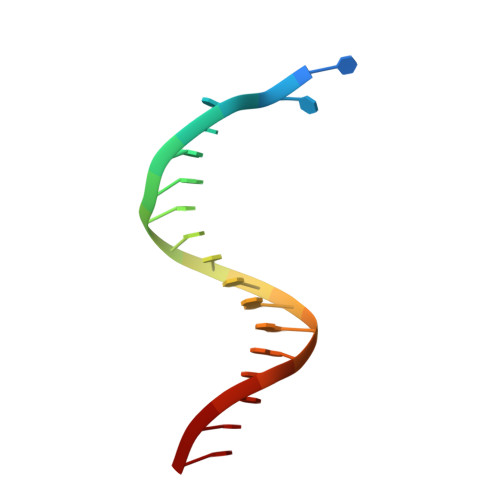
Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction
Kottur, J., Nair, D.T.(2018) Nucleic Acids Res 46: 5875-5885
- PubMed: 29850882
- DOI: https://doi.org/10.1093/nar/gky402
- Primary Citation of Related Structures:
5YUR, 5YUS, 5YUT, 5YUU, 5YUV, 5YUW, 5YUX, 5YUY, 5YUZ, 5YV0, 5YV1, 5YV2, 5YV3, 5YYD, 5YYE, 5ZLV, 6IG1 - PubMed Abstract:
DNA synthesis by DNA polymerases (dPols) is central to duplication and maintenance of the genome in all living organisms. dPols catalyze the formation of a phosphodiester bond between the incoming deoxynucleoside triphosphate and the terminal primer nucleotide with the release of a pyrophosphate (PPi) group. It is believed that formation of the phosphodiester bond is an endergonic reaction and PPi has to be hydrolyzed by accompanying pyrophosphatase enzymes to ensure that the free energy change of the DNA synthesis reaction is negative and it can proceed in the forward direction. The fact that DNA synthesis proceeds in vitro in the absence of pyrophosphatases represents a long-standing conundrum regarding the thermodynamics of the DNA synthesis reaction. Using time-resolved crystallography, we show that hydrolysis of PPi is an intrinsic and critical step of the DNA synthesis reaction catalyzed by dPols. The hydrolysis of PPi occurs after the formation of the phosphodiester bond and ensures that the DNA synthesis reaction is energetically favorable without the need for additional enzymes. Also, we observe that DNA synthesis is a two Mg2+ ion assisted stepwise associative SN2 reaction. Overall, this study provides deep temporal insight regarding the primary enzymatic reaction responsible for genome duplication.
- Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad 121 001, India.
Organizational Affiliation: