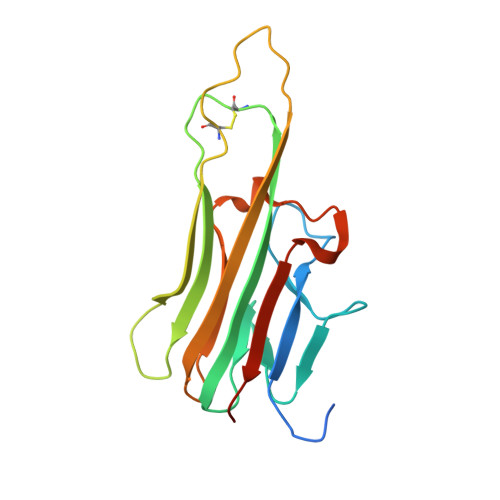
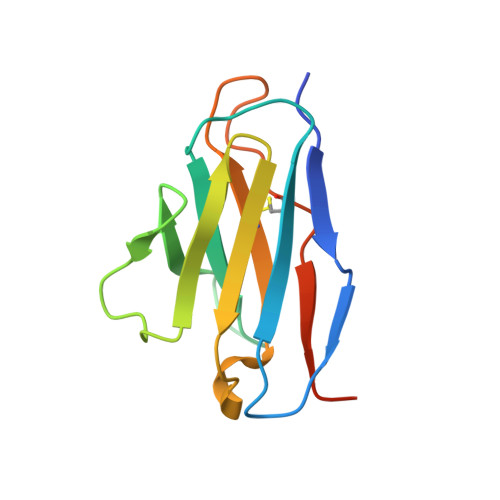
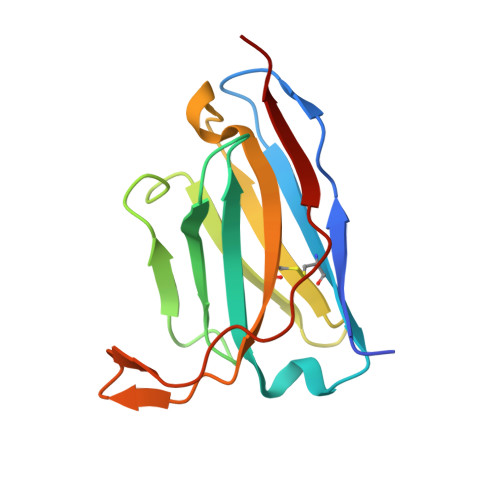
Structural basis for tumor necrosis factor blockade with the therapeutic antibody golimumab
Ono, M., Horita, S., Sato, Y., Nomura, Y., Iwata, S., Nomura, N.(2018) Protein Sci 27: 1038-1046
- PubMed: 29575262
- DOI: https://doi.org/10.1002/pro.3407
- Primary Citation of Related Structures:
5YOY - PubMed Abstract:
Tumor necrosis factor α (TNFα) is a proinflammatory cytokine, and elevated levels of TNFα in serum are associated with various autoimmune diseases, including rheumatoid arthritis (RA), ankylosing spondylitis (AS), Crohn's disease (CD), psoriasis, and systemic lupus erythaematosus. TNFα performs its pleiotropic functions by binding to two structurally distinct transmembrane receptors, TNF receptor (TNFR) 1 and TNFR2. Antibody-based therapeutic strategies that block excessive TNFα signaling have been shown to be effective in suppressing such harmful inflammatory conditions. Golimumab (Simponi®) is an FDA-approved fully human monoclonal antibody targeting TNFα that has been widely used for the treatment of RA, AS, and CD. However, the structural basis underlying the inhibitory action of golimumab remains unclear. Here, we report the crystal structure of the Fv fragment of golimumab in complex with TNFα at a resolution of 2.73 Å. The resolved structure reveals that golimumab binds to a distinct epitope on TNFα that does not overlap with the binding residues of TNFR2. Golimumab exerts its inhibitory effect by preventing binding of TNFR1 and TNFR2 to TNFα by steric hindrance. Golimumab does not induce conformational changes in TNFα that could affect receptor binding. This mode of action is specific to golimumab among the four anti-TNFα therapeutic antibodies currently approved for clinical use.
- Department of Cell Biology, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto, 606-8501, Japan.
Organizational Affiliation: