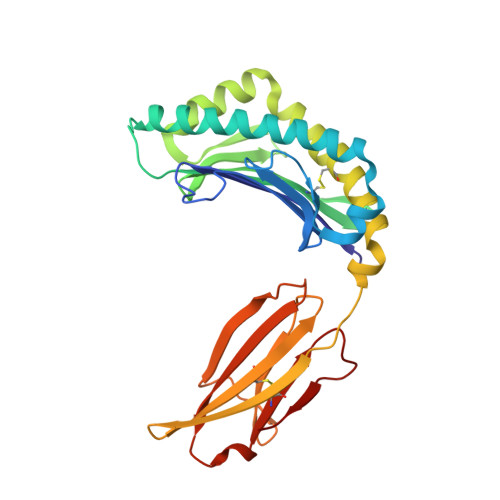
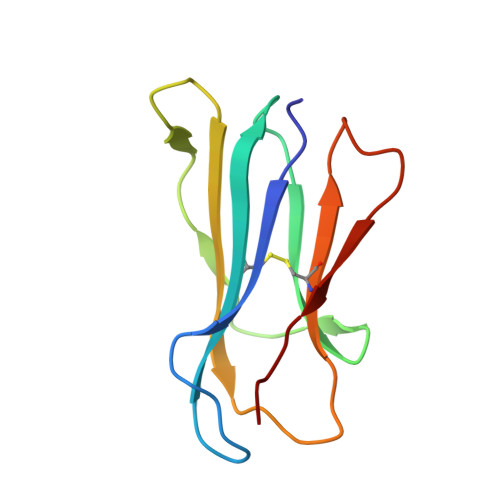
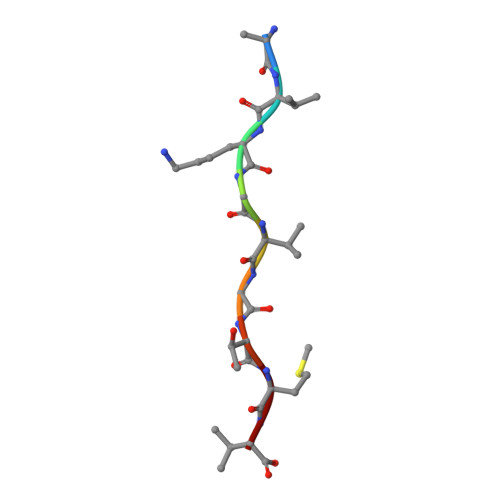
An Invariant Arginine in Common with MHC Class II Allows Extension at the C-Terminal End of Peptides Bound to Chicken MHC Class I.
Xiao, J., Xiang, W., Zhang, Y., Peng, W., Zhao, M., Niu, L., Chai, Y., Qi, J., Wang, F., Qi, P., Pan, C., Han, L., Wang, M., Kaufman, J., Gao, G.F., Liu, W.J.(2018) J Immunol 201: 3084-3095
- PubMed: 30341185
- DOI: https://doi.org/10.4049/jimmunol.1800611
- Primary Citation of Related Structures:
5YMV, 5YMW - PubMed Abstract:
MHC molecules are found in all jawed vertebrates and are known to present peptides to T lymphocytes. In mammals, peptides can hang out either end of the peptide-binding groove of classical class II molecules, whereas the N and C termini of peptides are typically tightly bound to specific pockets in classical class I molecules. The chicken MHC, like many nonmammalian vertebrates, has a single dominantly expressed classical class I molecule encoded by the BF2 locus. We determined the structures of BF2*1201 bound to two peptides and found that the C terminus of one peptide hangs outside of the groove with a conformation much like the peptides bound to class II molecules. We found that BF2*1201 binds many peptides that hang out of the groove at the C terminus, and the sequences and structures of this MHC class I allele were determined to investigate the basis for this phenomenon. The classical class I molecules of mammals have a nearly invariant Tyr (Tyr 84 in humans) that coordinates the peptide C terminus, but all classical class I molecules outside of mammals have an Arg in that position in common with mammalian class II molecules. We find that this invariant Arg residue switches conformation to allow peptides to hang out of the groove of BF2*1201, suggesting that this phenomenon is common in chickens and other nonmammalian vertebrates, perhaps allowing the single dominantly expressed class I molecule to bind a larger repertoire of peptides.
- Key Laboratory of Veterinary Bioproduction and Chemical Medicine of the Ministry of Agriculture, Engineering and Technology Research Center for Beijing Veterinary Peptide Vaccine Design and Preparation, Zhongmu Institutes of China Animal Husbandry Industry Co. Ltd., Beijing 100095, China.
Organizational Affiliation: