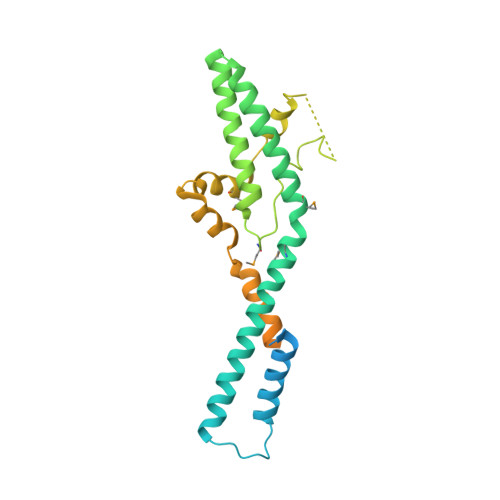
Structural and functional analysis of ribosome assembly factor Efg1.
Shu, S., Ye, K.(2018) Nucleic Acids Res 46: 2096-2106
- PubMed: 29361028
- DOI: https://doi.org/10.1093/nar/gky011
- Primary Citation of Related Structures:
5YMA - PubMed Abstract:
Ribosome biogenesis in eukaryotes is a complicated process that involves association and dissociation of numerous assembly factors and snoRNAs. The yeast small ribosomal subunit is first assembled into 90S pre-ribosomes in an ordered and dynamic manner. Efg1 is a protein with no recognizable domain that is associated with early 90S particles. Here, we determine the crystal structure of Efg1 from Chaetomium thermophilum at 3.3 Å resolution, revealing a novel elongated all-helical structure. Efg1 is not located in recently determined cryo-EM densities of 90S likely due to its low abundance in mature 90S. Genetic analysis in Saccharomyces cerevisiae shows that the functional core of Efg1 contains two helical hairpins composed of highly conserved residues. Depletion of Efg1 blocks 18S rRNA processing at sites A1 and A2, but not at site A0, and production of small ribosomal subunits. Efg1 is initially recruited by the 5' domain of 18S rRNA. Its absence disturbs the assembly of the 5' domain and inhibits release of U14 snoRNA from 90S. Our study shows that Efg1 is required for early assembly and reorganization of the 5' domain of 18S rRNA.
- Graduate School of Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing 100730, China.
Organizational Affiliation: