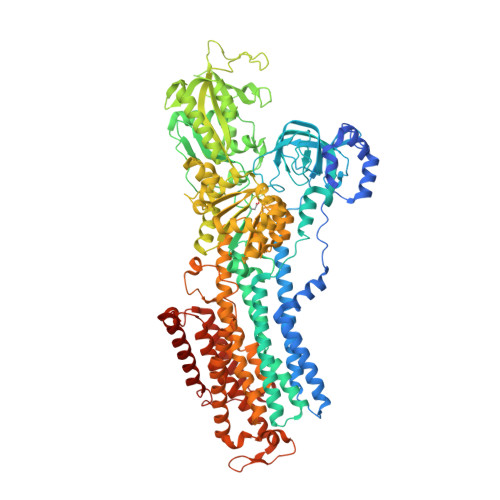
Crystal structures of the gastric proton pump
Abe, K., Irie, K., Nakanishi, H., Suzuki, H., Fujiyoshi, Y.(2018) Nature 556: 214-218
- PubMed: 29618813
- DOI: https://doi.org/10.1038/s41586-018-0003-8
- Primary Citation of Related Structures:
5YLU, 5YLV - PubMed Abstract:
The gastric proton pump-the H + , K + -ATPase-is a P-type ATPase responsible for acidifying the gastric juice down to pH 1. This corresponds to a million-fold proton gradient across the membrane of the parietal cell, the steepest known cation gradient of any mammalian tissue. The H + , K + -ATPase is an important target for drugs that treat gastric acid-related diseases. Here we present crystal structures of the H + , K + -ATPase in complex with two blockers, vonoprazan and SCH28080, in the luminal-open state, at 2.8 Å resolution. The drugs have partially overlapping but clearly distinct binding modes in the middle of a conduit running from the gastric lumen to the cation-binding site. The crystal structures suggest that the tight configuration at the cation-binding site lowers the pK a value of Glu820 sufficiently to enable the release of a proton even into the pH 1 environment of the stomach.
- Cellular and Structural Physiology Institute, Nagoya University, Nagoya, Japan. kabe@cespi.nagoya-u.ac.jp.
Organizational Affiliation: