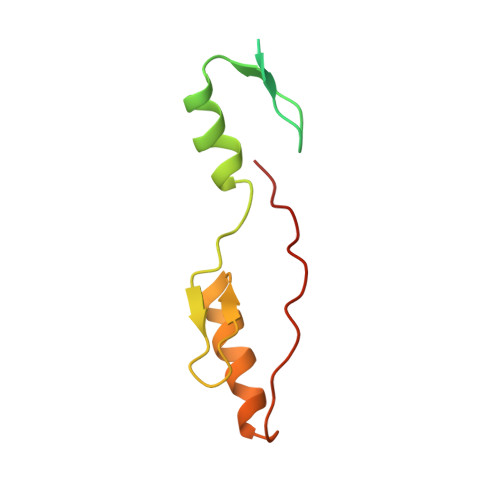
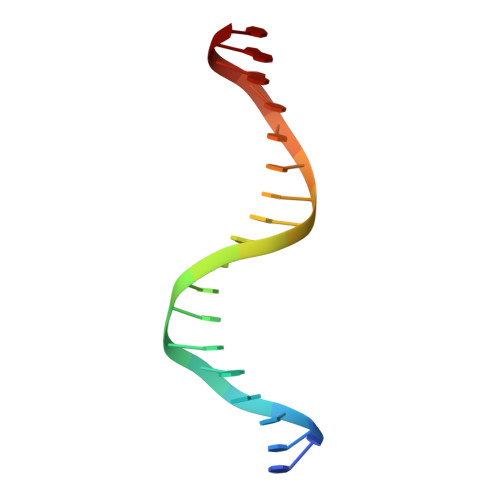
The 11th C2H2 zinc finger and an adjacent C-terminal arm are responsible for TZAP recognition of telomeric DNA.
Zhao, Y., Zhang, G., He, C., Mei, Y., Shi, Y., Li, F.(2018) Cell Res 28: 130-134
- PubMed: 29134956
- DOI: https://doi.org/10.1038/cr.2017.141
- Primary Citation of Related Structures:
5YJ3 - Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: