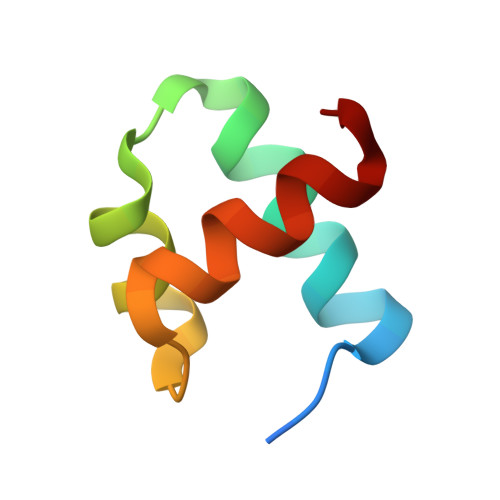
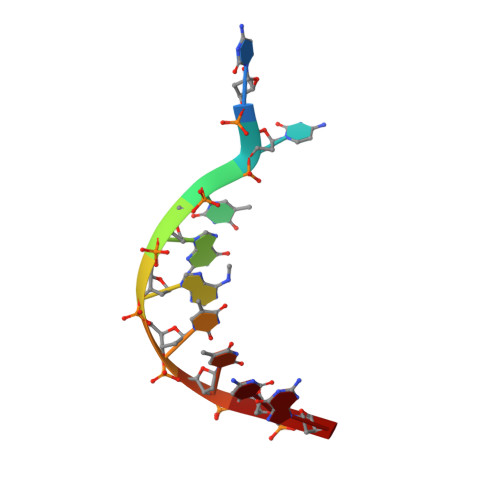
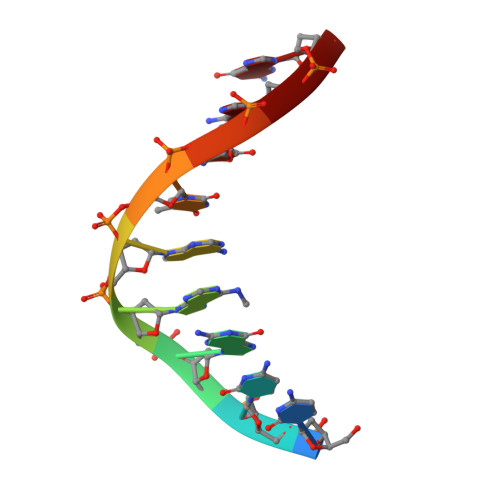
Structural insights into the unique mechanism of transcription activation by Caulobacter crescentus GcrA.
Wu, X., Haakonsen, D.L., Sanderlin, A.G., Liu, Y.J., Shen, L., Zhuang, N., Laub, M.T., Zhang, Y.(2018) Nucleic Acids Res 46: 3245-3256
- PubMed: 29514271
- DOI: https://doi.org/10.1093/nar/gky161
- Primary Citation of Related Structures:
5YIU, 5YIV, 5YIW, 5YIX, 5Z7I - PubMed Abstract:
Canonical bacterial transcription activators bind to non-transcribed promoter elements to increase transcription of their target genes. Here we report crystal structures of binary complexes comprising domains of Caulobacter crescentus GcrA, a noncanonical bacterial transcription factor, that support a novel mechanism for transcription activation through the preferential binding of methylated cis-regulatory elements and the promotion of open complex formation through an interaction with region 2 of the principal σ factor, σ70. We present crystal structures of the C-terminal, σ factor-interacting domain (GcrA-SID) in complex with domain 2 of σ70 (σ702), and the N-terminal, DNA-binding domain (GcrA-DBD) in complex with methylated double-stranded DNA (dsDNA). The structures reveal interactions essential for transcription activation and DNA recognition by GcrA. These structures, along with mutational analyses, support a mechanism of transcription activation in which GcrA associates with RNA polymerase (RNAP) prior to promoter binding through GcrA-SID, arming RNAP with a flexible GcrA-DBD. The RNAP-GcrA complex then binds and activates target promoters harboring a methylated GcrA binding site either upstream or downstream of the transcription start site.
- Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: