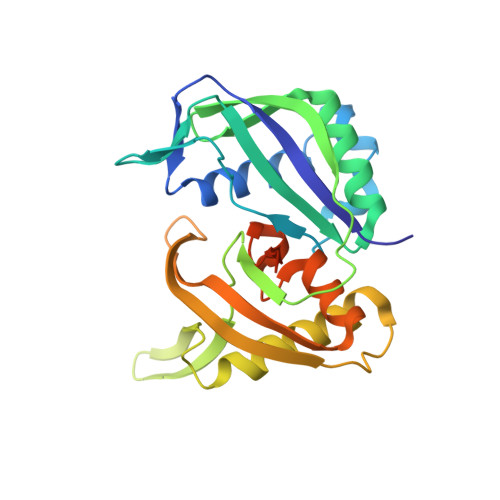
Expression, Purification, Crystallization, and X-ray Structural Analysis of CRISPR-Associated Protein Cas6 from Methanocaldococcus jannaschii
Lee, M.C., Tseng, S.T., Yang, J.C., Hsieh, T.J., Wu, S.C., Kuan, S.M., Chen, M.J., Chang, M.C., Wang, C.C., Chen, H.L., Fang, G.C., Huang, W.J., Ko, T.P., Chen, Y.(2018) Crystals (Basel) 7