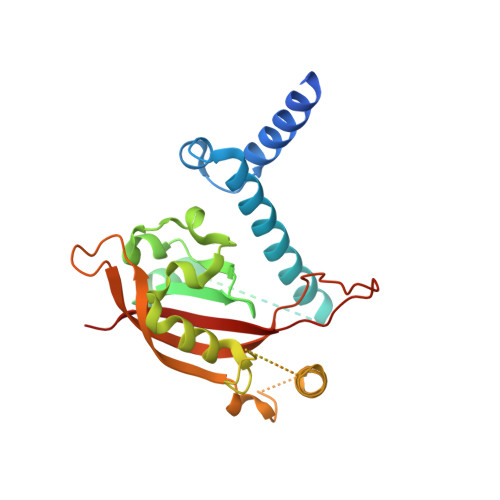
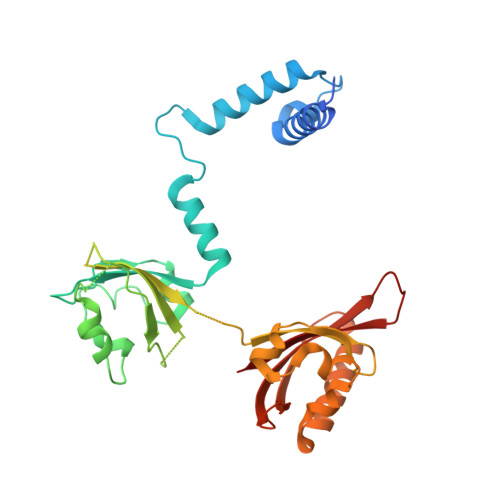
The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
Sakurai, S., Shimizu, T., Ohto, U.(2017) J Biological Chem 292: 17609-17616
- PubMed: 28904176
- DOI: https://doi.org/10.1074/jbc.M117.812974
- Primary Citation of Related Structures:
5Y7Y - PubMed Abstract:
2,3,7,8-Tetrachlorodibenzo- p -dioxin and related compounds are extraordinarily potent environmental toxic pollutants. Most of the 2,3,7,8-tetrachlorodibenzo- p -dioxin toxicities are mediated by aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor belonging to the basic helix-loop-helix (bHLH) Per-ARNT-Sim (PAS) family. Upon ligand binding, AhR forms a heterodimer with AhR nuclear translocator (ARNT) and induces the expression of genes involved in various biological responses. One of the genes induced by AhR encodes AhR repressor (AhRR), which also forms a heterodimer with ARNT and represses the activation of AhR-dependent transcription. The control of AhR activation is critical for managing AhR-mediated diseases, but the mechanisms by which AhRR represses AhR activation remain poorly understood, because of the lack of structural information. Here, we determined the structure of the AhRR-ARNT heterodimer by X-ray crystallography, which revealed an asymmetric intertwined domain organization presenting structural features that are both conserved and distinct among bHLH-PAS family members. The structures of AhRR-ARNT and AhR-ARNT were similar in the bHLH-PAS-A region, whereas the PAS-B of ARNT in the AhRR-ARNT complex exhibited a different domain arrangement in this family reported so far. The structure clearly disclosed that AhRR competitively represses AhR binding to ARNT and target DNA and further suggested the existence of an AhRR-ARNT-specific repression mechanism. This study provides a structural basis for understanding the mechanism by which AhRR represses AhR-mediated gene transcription.
- From the Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo 113-0033, Japan and.
Organizational Affiliation: