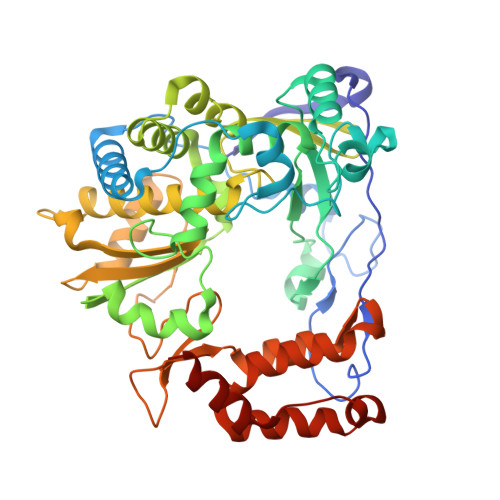
Crystal structure of the coxsackievirus A16 RNA-dependent RNA polymerase elongation complex reveals novel features in motif A dynamics
Bi, P., Shu, B., Gong, P.(2017) Virol Sin 32: 548-552
- PubMed: 29164396
- DOI: https://doi.org/10.1007/s12250-017-4066-8
- Primary Citation of Related Structures:
5Y6Z - PubMed Abstract:
The RNA-dependent RNA polymerases (RdRPs) encoded by RNA viruses represent a unique class of nucleic acid polymerases. Unlike other classes of single-subunit polymerases, viral RdRPs have evolved a unique conformational change in their palm domain to close the active site during catalysis. The hallmark of this conformational change is the backbone shift of the polymerase motif A from an "open" state to a "closed" state, allowing two universally conserved aspartic acid residues to orient toward each other for divalent metal binding and catalysis. The "closed" motif A conformation was only observed upon the binding of correct NTP in RdRP catalytic complexes or under rare conditions such as induced by a bound lutetium ion or a bound glutamate molecule. By solving the crystal structure of the catalytic elongation complex of the coxsackievirus RdRP, we in this work observed for the first time the "closed" motif A conformation in the absence of an NTP substrate or other conformational-change-inducing factors. This observation emphasizes the intrinsic dynamic features of viral RdRP motif A, and solidifies the structural basis for how this important structural element participates in catalytic events of the RdRPs.
- Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, 430071, China.
Organizational Affiliation: