Crystal structure of DAXX N-terminal four-helix bundle domain (4HB) in complex with ATRX
Huang, H., Patel, D.J.(2017) Nat Commun
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2017) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Death domain-associated protein 6,Transcriptional regulator ATRX | 119 | Homo sapiens | Mutation(s): 0 Gene Names: DAXX, BING2, DAP6, ATRX, RAD54L, XH2 EC: 3.6.4.12 | 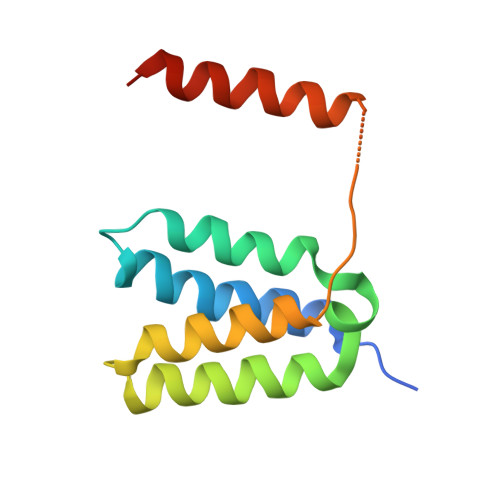 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P46100 (Homo sapiens) Explore P46100 Go to UniProtKB: P46100 | |||||
PHAROS: P46100 GTEx: ENSG00000085224 | |||||
Find proteins for Q9UER7 (Homo sapiens) Explore Q9UER7 Go to UniProtKB: Q9UER7 | |||||
PHAROS: Q9UER7 GTEx: ENSG00000204209 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P46100Q9UER7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.666 | α = 90 |
| b = 78.666 | β = 90 |
| c = 503.215 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data processing |
| pointless | data scaling |
| PHASER | phasing |