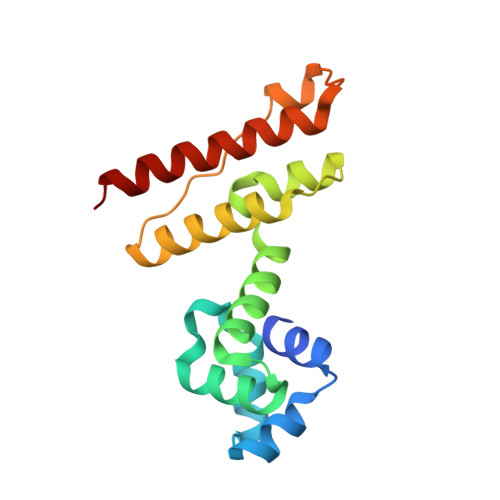
Crystal structure of an anti-CRISPR protein, AcrIIA1
Ka, D., An, S.Y., Suh, J.Y., Bae, E.(2018) Nucleic Acids Res 46: 485-492
- PubMed: 29182776
- DOI: https://doi.org/10.1093/nar/gkx1181
- Primary Citation of Related Structures:
5Y69, 5Y6A - PubMed Abstract:
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins provide bacteria with RNA-based adaptive immunity against phage infection. To counteract this defense mechanism, phages evolved anti-CRISPR (Acr) proteins that inactivate the CRISPR-Cas systems. AcrIIA1, encoded by Listeria monocytogenes prophages, is the most prevalent among the Acr proteins targeting type II-A CRISPR-Cas systems and has been used as a marker to identify other Acr proteins. Here, we report the crystal structure of AcrIIA1 and its RNA-binding affinity. AcrIIA1 forms a dimer with a novel two helical-domain architecture. The N-terminal domain of AcrIIA1 exhibits a helix-turn-helix motif similar to transcriptional factors. When overexpressed in Escherichia coli, AcrIIA1 associates with RNAs, suggesting that AcrIIA1 functions via nucleic acid recognition. Taken together, the unique structural and functional features of AcrIIA1 suggest its distinct mode of Acr activity, expanding the diversity of the inhibitory mechanisms employed by Acr proteins.
- Department of Agricultural Biotechnology, Seoul National University, Seoul 08826, Korea.
Organizational Affiliation: