Structural view of the helicase reveals that Zika virus uses a conserved mechanism for unwinding RNA
Li, L., Wang, J., Jia, Z., Shaw, N.(2018) Acta Crystallogr F Struct Biol Commun 74: 205-213
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NS3 helicase | 440 | Zika virus | Mutation(s): 0 EC: 3.4.21.91 (PDB Primary Data), 3.6.1.15 (PDB Primary Data), 3.6.4.13 (PDB Primary Data) | 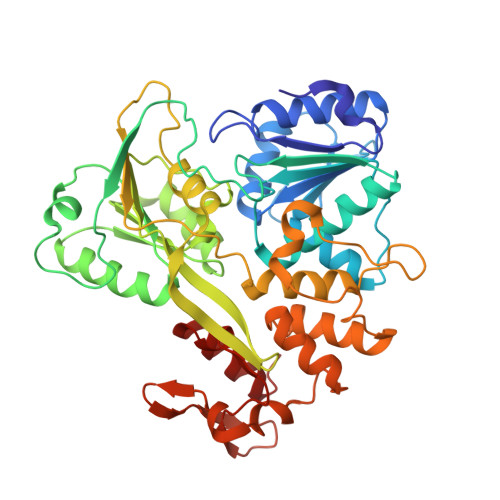 | |
UniProt | |||||
Find proteins for Q32ZE1 (Zika virus) Explore Q32ZE1 Go to UniProtKB: Q32ZE1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q32ZE1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP Query on ANP | B [auth A] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| MN (Subject of Investigation/LOI) Query on MN | C [auth A] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.754 | α = 90 |
| b = 68.779 | β = 92.5 |
| c = 56.971 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data processing |
| PHENIX | phasing |