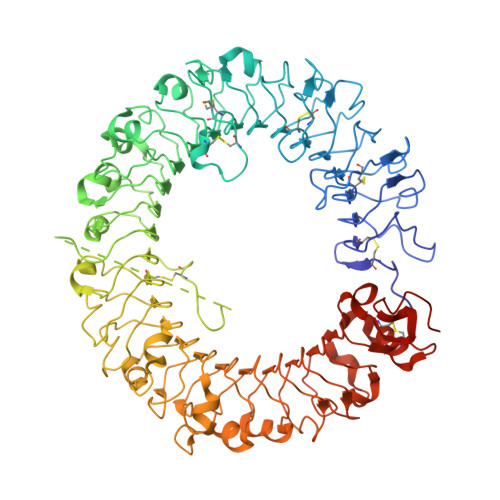
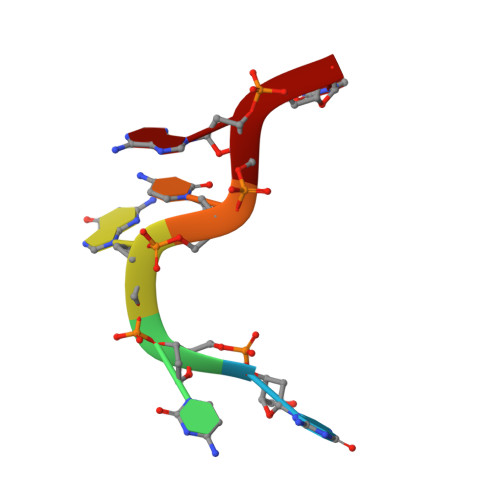
Toll-like Receptor 9 Contains Two DNA Binding Sites that Function Cooperatively to Promote Receptor Dimerization and Activation
Ohto, U., Ishida, H., Shibata, T., Sato, R., Miyake, K., Shimizu, T.(2018) Immunity 48: 649-658.e4
- PubMed: 29625894
- DOI: https://doi.org/10.1016/j.immuni.2018.03.013
- Primary Citation of Related Structures:
5Y3J, 5Y3K, 5Y3L, 5Y3M - PubMed Abstract:
Toll-like receptor 9 (TLR9) recognizes DNA containing CpG motifs derived from bacteria and viruses and activates the innate immune response to eliminate them. TLR9 is known to bind to CpG DNA, and here, we identified another DNA binding site in TLR9 that binds DNA containing cytosine at the second position from the 5' end (5'-xCx DNA). 5'-xCx DNAs bound to TLR9 in the presence of CpG DNA and cooperatively promoted dimerization and activation of TLR9. Binding at both sites was important for efficient activation of TLR9. The 5'-xCx DNA bound the site corresponding to the nucleoside binding site in TLR7 and TLR8 as revealed by the structural analysis. This study revealed that TLR9 recognizes two types of DNA through its two binding sites for efficient activation. This information may contribute to the development of drugs that control the activity of TLR9.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Hongo, Bunkyo-ku, Tokyo 113-0033, Japan. Electronic address: umeji@mol.f.u-tokyo.ac.jp.
Organizational Affiliation: