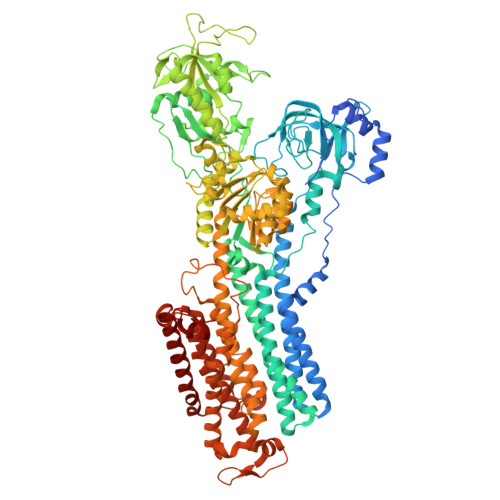
The cryo-EM structure of gastric H(+),K(+)-ATPase with bound BYK99, a high-affinity member of K(+)-competitive, imidazo[1,2-a]pyridine inhibitors
Abe, K., Shimokawa, J., Naito, M., Munson, K., Vagin, O., Sachs, G., Suzuki, H., Tani, K., Fujiyoshi, Y.(2017) Sci Rep 7: 6632-6632
- PubMed: 28747707
- DOI: https://doi.org/10.1038/s41598-017-06698-8
- Primary Citation of Related Structures:
5Y0B - PubMed Abstract:
The gastric proton pump H + ,K + -ATPase acidifies the gastric lumen, and thus its inhibitors, including the imidazo[1,2-a]pyridine class of K + -competitive acid blockers (P-CABs), have potential application as acid-suppressing drugs. We determined the electron crystallographic structure of H + ,K + -ATPase at 6.5 Å resolution in the E2P state with bound BYK99, a potent P-CAB with a restricted ring structure. The BYK99 bound structure has an almost identical profile to that of a previously determined structure with bound SCH28080, the original P-CAB prototype, but is significantly different from the previously reported P-CAB-free form, illustrating a common conformational change is required for P-CAB binding. The shared conformational changes include a distinct movement of transmembrane helix 2 (M2), from its position in the previously reported P-CAB-free form, to a location proximal to the P-CAB binding site in the present BYK99-bound structure. Site-specific mutagenesis within M2 revealed that D137 and N138, which face the P-CAB binding site in our model, significantly affect the inhibition constant (K i ) of P-CABs. We also found that A335 is likely to be near the bridging nitrogen at the restricted ring structure of the BYK99 inhibitor. These provide clues to elucidate the binding site parameters and mechanism of P-CAB inhibition of gastric acid secretion.
- Graduate School of Pharmaceutical Sciences, Nagoya University, Nagoya, 464-8601, Japan. kabe@cespi.nagoya-u.ac.jp.
Organizational Affiliation: