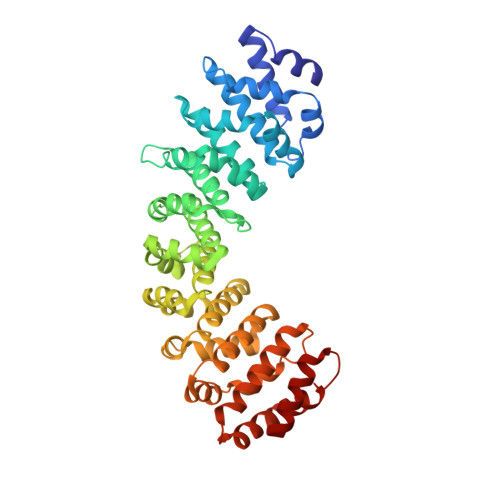
Crystal structure of importin-alpha 3 bound to the nuclear localization signal of Ran-binding protein 3
Koyama, M., Matsuura, Y.(2017) Biochem Biophys Res Commun 491: 609-613
- PubMed: 28760339
- DOI: https://doi.org/10.1016/j.bbrc.2017.07.155
- Primary Citation of Related Structures:
5XZX - PubMed Abstract:
Ran-binding protein 3 (RanBP3) is a primarily nuclear Ran-binding protein that functions as an accessory factor in the Ran GTPase system. RanBP3 associates with Ran-specific nucleotide exchange factor RCC1 and enhances its catalytic activity towards Ran. RanBP3 also promotes CRM1-mediated nuclear export as well as CRM1-independent nuclear export of β-catenin, Smad2, and Smad3. Nuclear import of RanBP3 is dependent on the nuclear import adaptor protein importin-α and, RanBP3 is imported more efficiently by importin-α3 than by other members of the importin-α family. Protein kinase signaling pathways control nucleocytoplasmic transport through phosphorylation of RanBP3 at Ser58, immediately C-terminal to the nuclear localization signal (NLS) in the N-terminal region of RanBP3. Here we report the crystal structure of human importin-α3 bound to an N-terminal fragment of human RanBP3 containing the NLS sequence that is necessary and sufficient for nuclear import. The structure reveals that RanBP3 binds to importin-α3 residues that are strictly conserved in all seven isoforms of human importin-α at the major NLS-binding site, indicating that the region of importin-α outside the NLS-binding site, possibly the autoinhibotory importin-β1-binding domain, may be the key determinant for the preferential binding of RanBP3 to importin-α3. Computational docking simulation indicates that phosphorylation of RanBP3 at Ser58 could potentially stabilize the association of RanBP3 with importin-α through interactions between the phosphate moiety of phospho-Ser58 of RanBP3 and a cluster of basic residues (Arg96 and Lys97 in importin-α3) on armadillo repeat 1 of importin-α.
- Division of Biological Science, Graduate School of Science, Nagoya University, Japan.
Organizational Affiliation: