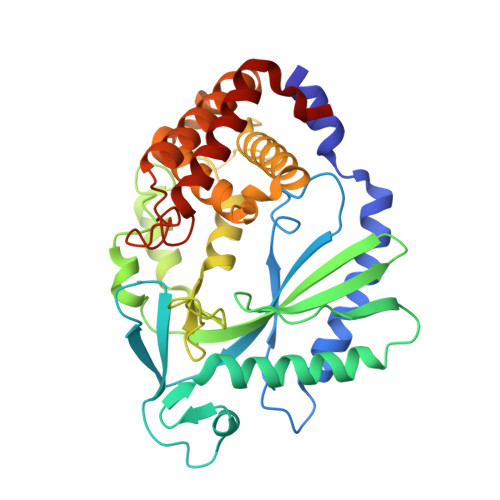
Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Vincent, J., Adura, C., Gao, P., Luz, A., Lama, L., Asano, Y., Okamoto, R., Imaeda, T., Aida, J., Rothamel, K., Gogakos, T., Steinberg, J., Reasoner, S., Aso, K., Tuschl, T., Patel, D.J., Glickman, J.F., Ascano, M.(2017) Nat Commun 8: 750-750
- PubMed: 28963528
- DOI: https://doi.org/10.1038/s41467-017-00833-9
- Primary Citation of Related Structures:
5XZB, 5XZE, 5XZG - PubMed Abstract:
Cyclic GMP-AMP synthase is essential for innate immunity against infection and cellular damage, serving as a sensor of DNA from pathogens or mislocalized self-DNA. Upon binding double-stranded DNA, cyclic GMP-AMP synthase synthesizes a cyclic dinucleotide that initiates an inflammatory cellular response. Mouse studies that recapitulate causative mutations in the autoimmune disease Aicardi-Goutières syndrome demonstrate that ablating the cyclic GMP-AMP synthase gene abolishes the deleterious phenotype. Here, we report the discovery of a class of cyclic GMP-AMP synthase inhibitors identified by a high-throughput screen. These compounds possess defined structure-activity relationships and we present crystal structures of cyclic GMP-AMP synthase, double-stranded DNA, and inhibitors within the enzymatic active site. We find that a chemically improved member, RU.521, is active and selective in cellular assays of cyclic GMP-AMP synthase-mediated signaling and reduces constitutive expression of interferon in macrophages from a mouse model of Aicardi-Goutières syndrome. RU.521 will be useful toward understanding the biological roles of cyclic GMP-AMP synthase and can serve as a molecular scaffold for development of future autoimmune therapies.Upon DNA binding cyclic GMP-AMP synthase (cGAS) produces a cyclic dinucleotide, which leads to the upregulation of inflammatory genes. Here the authors develop small molecule cGAS inhibitors, functionally characterize them and present the inhibitor and DNA bound cGAS crystal structures, which will facilitate drug development.
- Vanderbilt University School of Medicine, Nashville, TN, 37027, USA.
Organizational Affiliation: