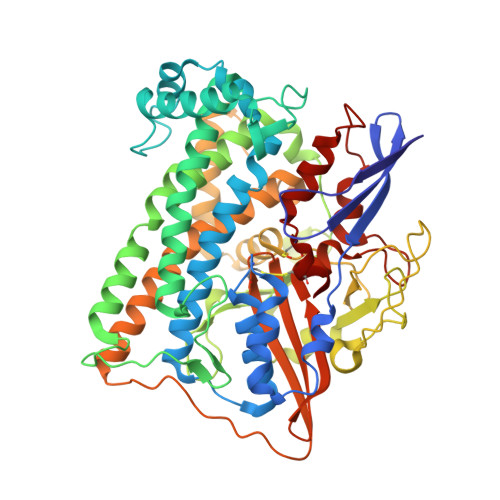
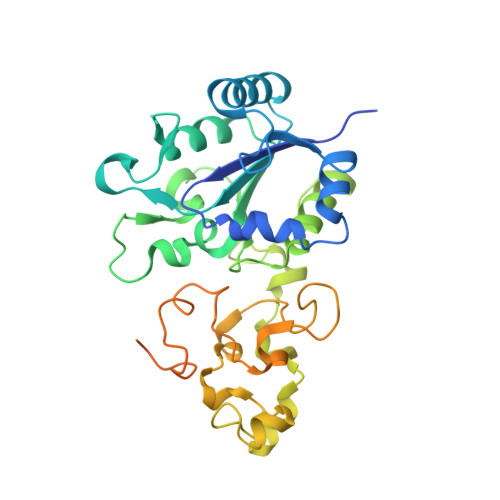
Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Noor, N.D.M., Matsuura, H., Nishikawa, K., Tai, H., Hirota, S., Kim, J., Kang, J., Tateno, M., Yoon, K.S., Ogo, S., Kubota, S., Shomura, Y., Higuchi, Y.(2018) Chem Commun (Camb) 54: 12385-12388
- PubMed: 30328414
- DOI: https://doi.org/10.1039/c8cc06261g
- Primary Citation of Related Structures:
5XVB, 5XVC, 5XVD - PubMed Abstract:
Citrobacter sp. S-77 [NiFe]-hydrogenase harbors a standard [4Fe-4S] cluster proximal to the Ni-Fe active site. The presence of relocatable water molecules and a flexible aspartate enables the [4Fe-4S] to display redox-dependent conformational changes. These structural features are proposed to be the key aspects that protect the active site from O2 attack.
- Department of Life Science, Graduate School of Life Science, University of Hyogo, 3-2-1 Koto, Kamigori-cho, Ako-gun, Hyogo 678-1297, Japan. hig@sci.u-hyogo.ac.jp.
Organizational Affiliation: