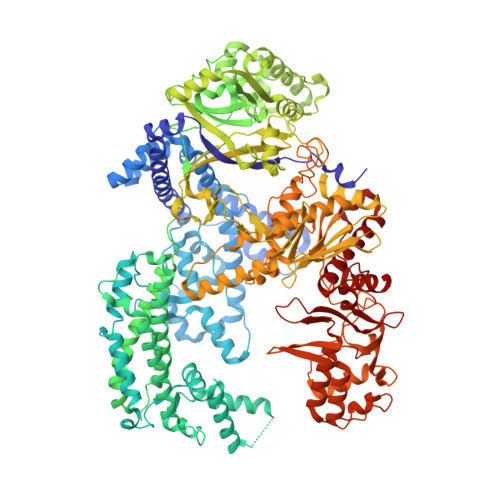
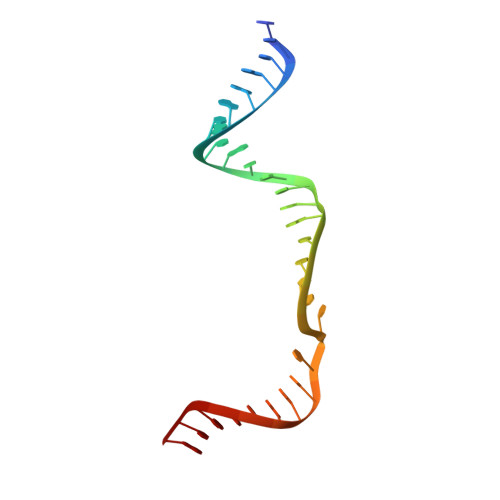
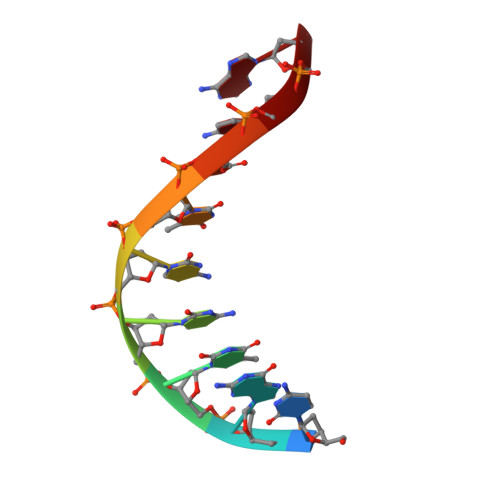
Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Yamano, T., Zetsche, B., Ishitani, R., Zhang, F., Nishimasu, H., Nureki, O.(2017) Mol Cell 67: 633-645.e3
- PubMed: 28781234
- DOI: https://doi.org/10.1016/j.molcel.2017.06.035
- Primary Citation of Related Structures:
5XUS, 5XUT, 5XUU, 5XUZ - PubMed Abstract:
The RNA-guided Cpf1 (also known as Cas12a) nuclease associates with a CRISPR RNA (crRNA) and cleaves the double-stranded DNA target complementary to the crRNA guide. The two Cpf1 orthologs from Acidaminococcus sp. (AsCpf1) and Lachnospiraceae bacterium (LbCpf1) have been harnessed for eukaryotic genome editing. Cpf1 requires a specific nucleotide sequence, called a protospacer adjacent motif (PAM), for target recognition. Besides the canonical TTTV PAM, Cpf1 recognizes suboptimal C-containing PAMs. Here, we report four crystal structures of LbCpf1 in complex with the crRNA and its target DNA containing either TTTA, TCTA, TCCA, or CCCA as the PAM. These structures revealed that, depending on the PAM sequences, LbCpf1 undergoes conformational changes to form altered interactions with the PAM-containing DNA duplexes, thereby achieving the relaxed PAM recognition. Collectively, the present structures advance our mechanistic understanding of the PAM-dependent, crRNA-guided DNA cleavage by the Cpf1 family nucleases.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-0032, Japan.
Organizational Affiliation: