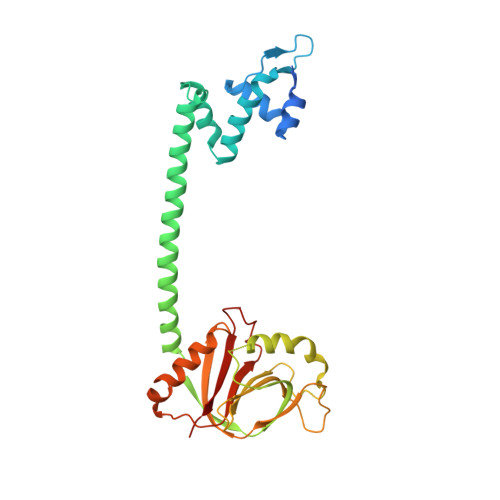
Crystal structure of BrlR with c-di-GMP
Raju, H., Sharma, R.(2017) Biochem Biophys Res Commun 490: 260-264
- PubMed: 28619510
- DOI: https://doi.org/10.1016/j.bbrc.2017.06.033
- Primary Citation of Related Structures:
5XQL - PubMed Abstract:
The transcriptional regulator BrlR is a member of the MerR family of multidrug transport activators in Pseudomonas aeruginosa. Recent study indicates that BrlR is a novel 3',5'-cyclic diguanylic acid (c-di-GMP) receptor and can be activated by c-di-GMP. To gain insight into BrlR function, we determined the structure of BrlR with c-di-GMP complex structure to 2.5 Å. The structure and size exclusion chromatography (SEC) data revealed BrlR forms a tetramer and each BrlR protomer consists of three parts, DNA-binding domain, a coiled-coil region and GyrI-like domain. There are two different c-di-GMP binding sites located mainly at the DNA binding domain of each BrlR protomer and do not overlap with the GyrI-like domain. The drug-binding pocket in GyrI-like domain is much conserved indicating it can also bind flat-shaped molecules like other multidrug resistance (MDR) proteins.
- Department of Biophysics, Molecular Biology and Bioinformatics, University of Calcutta, Kolkata, 700009, West Bengal, India.
Organizational Affiliation: