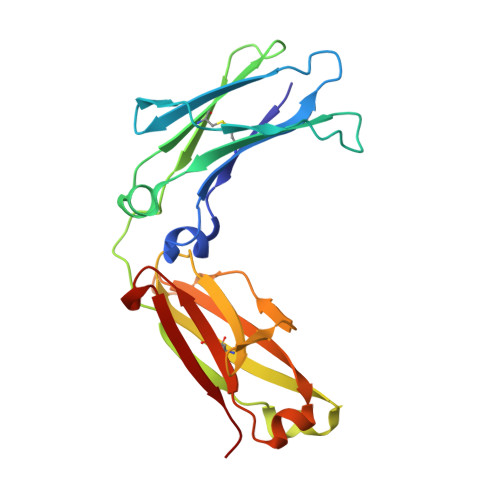
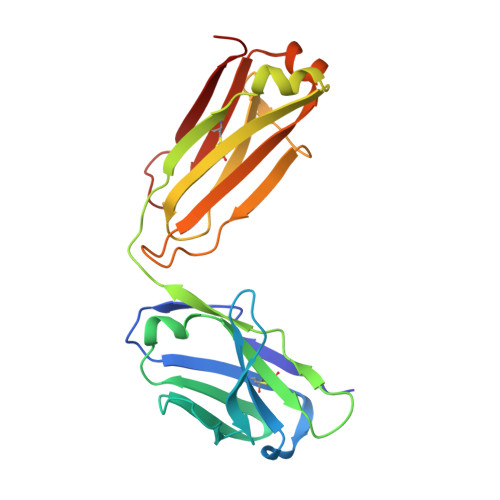
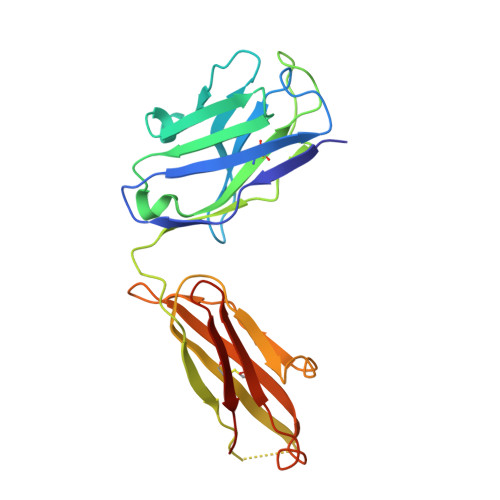
Structure-function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline-encoded antibody autoreactivity.
Shiroishi, M., Ito, Y., Shimokawa, K., Lee, J.M., Kusakabe, T., Ueda, T.(2018) J Biological Chem 293: 7008-7016
- PubMed: 29523691
- DOI: https://doi.org/10.1074/jbc.M117.814475
- Primary Citation of Related Structures:
5XMH - PubMed Abstract:
Rheumatoid factors (RFs) are autoantibodies against the fragment-crystallizable (Fc) region of IgG. In individuals with hematological diseases such as cryoglobulinemia and certain B cell lymphoma forms, the RFs derived from specific heavy- and light-chain germline pairs, so-called "stereotypic RFs," are frequently produced in copious amounts and form immune complexes with IgG in serum. Of note, many structural details of the antigen recognition mechanisms in RFs are unclear. Here we report the crystal structure of the RF YES8c derived from the IGHV1-69 / IGKV3-20 germline pair, the most common of the stereotypic RFs, in complex with human IgG1-Fc at 2.8 Å resolution. We observed that YES8c binds to the CH2-CH3 elbow in the canonical antigen-binding manner involving a large antigen-antibody interface. On the basis of this observation, combined with mutational analyses, we propose a recognition mechanism common to IGHV1-69/IGKV3-20 RFs: (1) the interaction of the Leu 432 -His 435 region of Fc enables the highly variable complementarity-determining region (CDR)-H3 to participate in the binding, (2) the hydrophobic tip in the CDR-H2 typical of IGHV1-69 antibodies recognizes the hydrophobic patch on Fc, and (3) the interaction of the highly conserved RF light chain with Fc is important for RF activity. These features may determine the putative epitope common to the IGHV1-69/IGKV3-20 RFs. We also showed that some mutations in the binding site of RF increase the affinity to Fc, which may aggravate hematological diseases. Our findings unravel the structural basis for germline-encoded antibody autoreactivity.
- From the Laboratory of Protein Structure, Function, and Design, Graduate School of Pharmaceutical Sciences, Kyushu University, Fukuoka 812-8582, Japan and shiroish@phar.kyushu-u.ac.jp.
Organizational Affiliation: