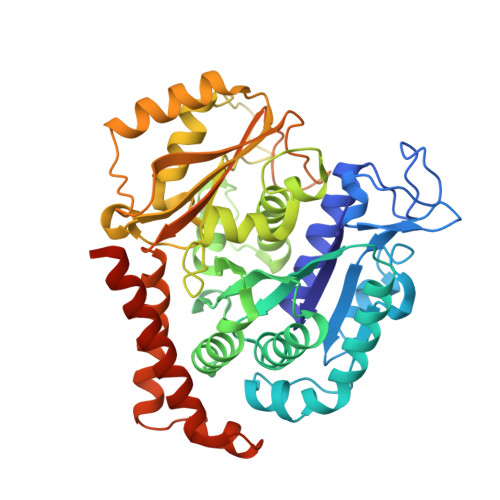
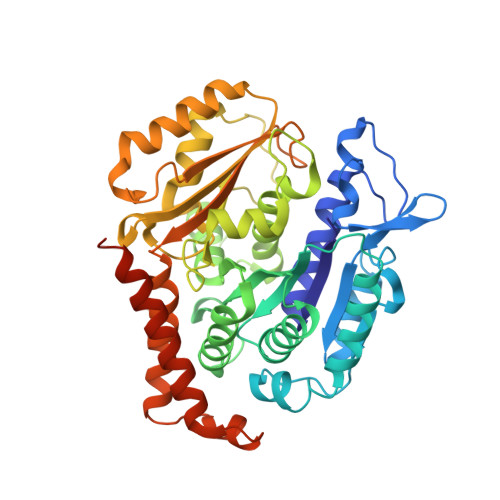
Structure of 4'-demethylepipodophyllotoxin in complex with tubulin provides a rationale for drug design
Niu, L., Wang, Y., Wang, C., Wang, Y., Jiang, X., Ma, L., Wu, C., Yu, Y., Chen, Q.(2017) Biochem Biophys Res Commun 493: 718-722
- PubMed: 28864414
- DOI: https://doi.org/10.1016/j.bbrc.2017.08.125
- Primary Citation of Related Structures:
5XLT - PubMed Abstract:
Microtubules consists of αβ-tubulin heterodimers and are highly attractive targets for anti-cancer drugs. A broad range of agents have been identified to bind to tubulin and interfere with microtubule assembly, including colchicine binding site inhibitors (CBSIs). Podophyllotoxin is a CBSI that inhibits the assembly of microtubules. However, for a long time, the design and development of podophyllotoxin family drugs have been hindered by the lack of high-resolution structural information of the tubulin-agent complex. We report the first high-resolution (2.8 Å) structure of a podophyllotoxin family agent (4'-demethylepipodophyllotoxin, DMEP) complexed with tubulin and revealed the detailed interactions between DMEP and tubulin. Comparison of this structure and other CBSIs explains previous results of the structure-activity-relationship (SAR) studies, and provides insights into the development of new podophyllotoxin derivatives targeting the colchicine site.
- Cancer Center, West China Hospital, Sichuan University, Collaborative Innovation Center of Biotherapy, Chengdu, 610041, PR China.
Organizational Affiliation: