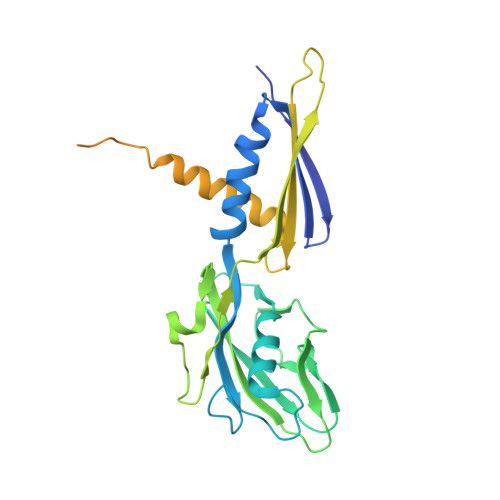
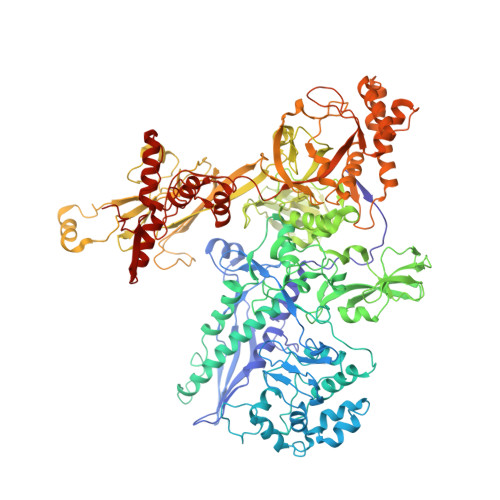
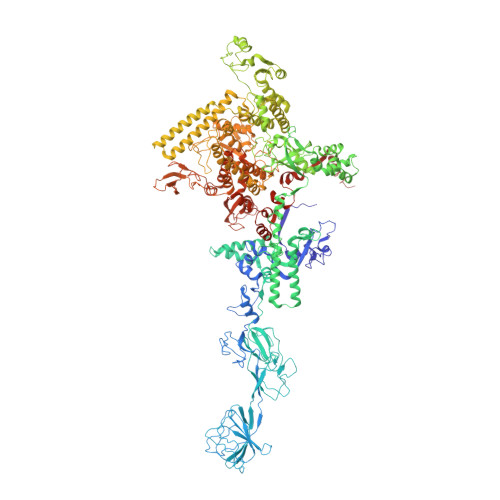
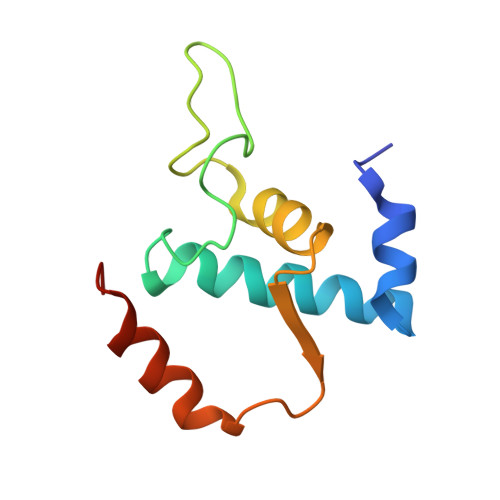
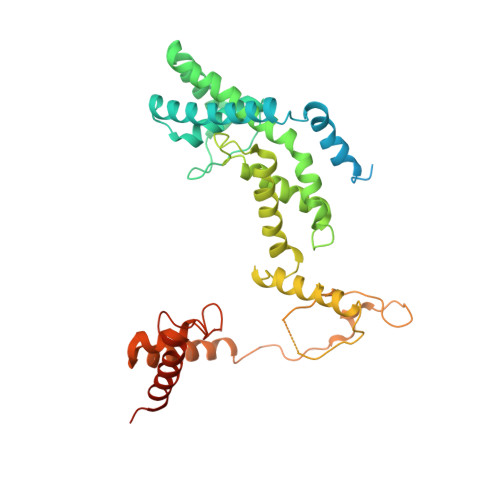
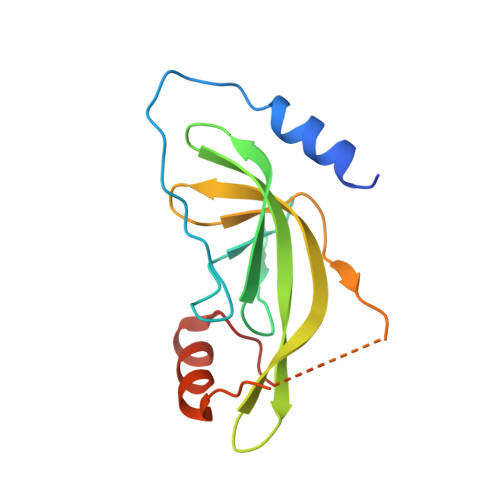
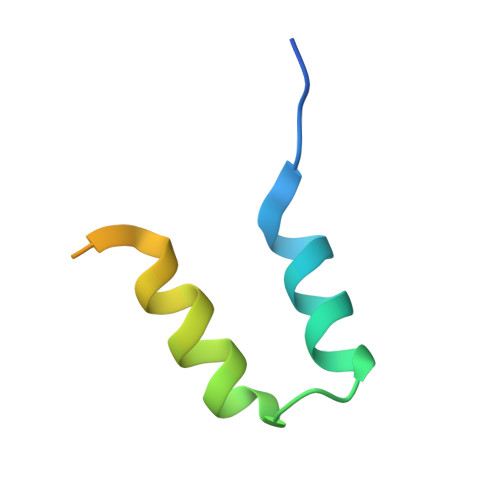
A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation
Ooi, W.Y., Murayama, Y., Mekler, V., Minakhin, L., Severinov, K., Yokoyama, S., Sekine, S.I.(2018) Nucleic Acids Res 46: 431-441
- PubMed: 29165680
- DOI: https://doi.org/10.1093/nar/gkx1162
- Primary Citation of Related Structures:
5XJ0 - PubMed Abstract:
RNA polymerase (RNAP) is a major target of gene regulation. Thermus thermophilus bacteriophage P23-45 encodes two RNAP binding proteins, gp39 and gp76, which shut off host gene transcription while allowing orderly transcription of phage genes. We previously reported the structure of the T. thermophilus RNAP•σA holoenzyme complexed with gp39. Here, we solved the structure of the RNAP•σA holoenzyme bound with both gp39 and gp76, which revealed an unprecedented inhibition mechanism by gp76. The acidic protein gp76 binds within the RNAP cleft and occupies the path of the template DNA strand at positions -11 to -4, relative to the transcription start site at +1. Thus, gp76 obstructs the formation of an open promoter complex and prevents transcription by T. thermophilus RNAP from most host promoters. gp76 is less inhibitory for phage transcription, as tighter RNAP interaction with the phage promoters allows the template DNA to compete with gp76 for the common binding site. gp76 also inhibits Escherichia coli RNAP highlighting the template-DNA binding site as a new target site for developing antibacterial agents.
- RIKEN Center for Life Science Technologies, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan.
Organizational Affiliation: