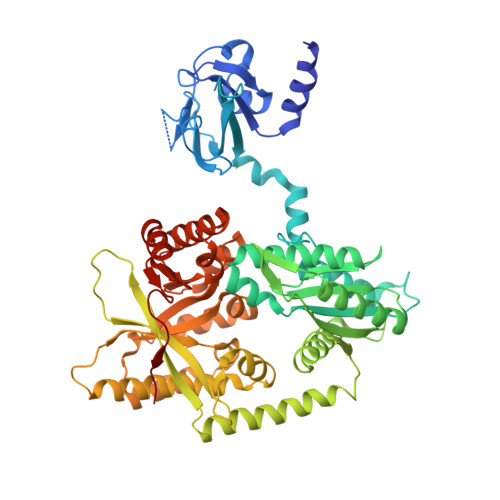
Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
Liu, C., Liew, C.W., Wong, Y.H., Tan, S.T., Poh, W.H., Manimekalai, M.S.S., Rajan, S., Xin, L., Liang, Z.X., Gruber, G., Rice, S.A., Lescar, J.(2018) J Bacteriol 200
- PubMed: 29109186
- DOI: https://doi.org/10.1128/JB.00515-17
- Primary Citation of Related Structures:
5XGB, 5XGD, 5XGE - PubMed Abstract:
RbdA is a positive r egulator of b iofilm d ispersal of Pseudomonas aeruginosa Its cytoplasmic region (cRbdA) comprises an N-terminal Per-ARNT-Sim (PAS) domain followed by a diguanylate cyclase (GGDEF) domain and an EAL domain, whose phosphodiesterase activity is allosterically stimulated by GTP binding to the GGDEF domain. We report crystal structures of cRbdA and of two binary complexes: one with GTP/Mg 2+ bound to the GGDEF active site and one with the EAL domain bound to the c-di-GMP substrate. These structures unveil a 2-fold symmetric dimer stabilized by a closely packed N-terminal PAS domain and a noncanonical EAL dimer. The autoinhibitory switch is formed by an α-helix (S-helix) immediately N-terminal to the GGDEF domain that interacts with the EAL dimerization helix (α 6-E ) of the other EAL monomer and maintains the protein in a locked conformation. We propose that local conformational changes in cRbdA upon GTP binding lead to a structure with the PAS domain and S-helix shifted away from the GGDEF-EAL domains, as suggested by small-angle X-ray scattering (SAXS) experiments. Domain reorientation should be facilitated by the presence of an α-helical lever (H-helix) that tethers the GGDEF and EAL regions, allowing the EAL domain to rearrange into an active dimeric conformation. IMPORTANCE Biofilm formation by bacterial pathogens increases resistance to antibiotics. RbdA positively regulates biofilm dispersal of Pseudomonas aeruginosa The crystal structures of the cytoplasmic region of the RbdA protein presented here reveal that two evolutionarily conserved helices play an important role in regulating the activity of RbdA, with implications for other GGDEF-EAL dual domains that are abundant in the proteomes of several bacterial pathogens. Thus, this work may assist in the development of small molecules that promote bacterial biofilm dispersal.
- School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.
Organizational Affiliation: