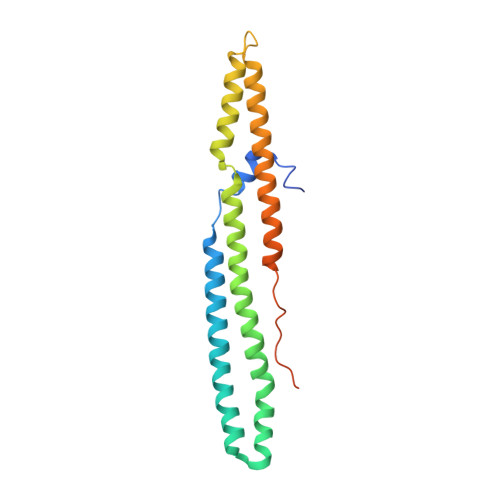
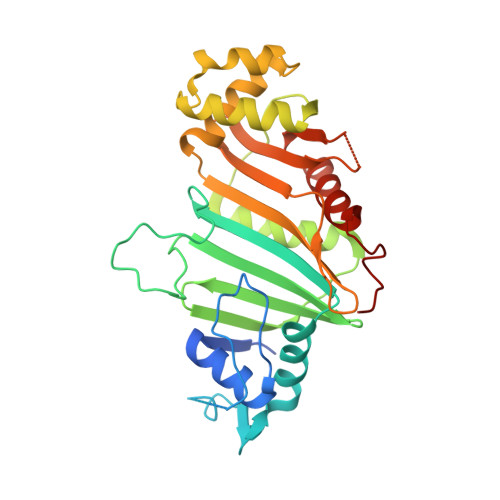
Structural basis of the PE-PPE protein interaction in Mycobacterium tuberculosis.
Chen, X., Cheng, H.F., Zhou, J., Chan, C.Y., Lau, K.F., Tsui, S.K., Au, S.W.(2017) J Biological Chem 292: 16880-16890
- PubMed: 28842489
- DOI: https://doi.org/10.1074/jbc.M117.802645
- Primary Citation of Related Structures:
5XFS - PubMed Abstract:
Mycobacterium tuberculosis ( Mtb ), the causative agent of tuberculosis, has developed multiple strategies to adapt to the human host. The five type VII secretion systems, ESX-1-5, direct the export of many virulence-promoting protein effectors across the complex mycobacterial cell wall. One class of ESX substrates is the PE-PPE family of proteins, which is unique to mycobacteria and essential for infection, antigenic variation, and host-pathogen interactions. The genome of Mtb encodes 168 PE-PPE proteins. Many of them are thought to be secreted through ESX-5 secretion system and to function in pairs. However, understanding of the specific pairing of PE-PPE proteins and their structure-function relationship is limited by the challenging purification of many PE-PPE proteins, and our knowledge of the PE-PPE interactions therefore has been restricted to the PE25-PPE41 pair and its complex with the ESX-5 secretion system chaperone EspG5. Here, we report the crystal structure of a new PE-PPE pair, PE8-PPE15, in complex with EspG5. Our structure revealed that the EspG5-binding sites on PPE15 are relatively conserved among Mtb PPE proteins, suggesting that EspG5-PPE15 represents a more typical model for EspG5-PPE interactions than EspG5-PPE41. A structural comparison with the PE25-PPE41 complex disclosed conformational changes in the four-helix bundle structure and a unique binding mode in the PE8-PPE15 pair. Moreover, homology-modeling and mutagenesis studies further delineated the molecular determinants of the specific PE-PPE interactions. These findings help develop an atomic algorithm of ESX-5 substrate recognition and PE-PPE pairing.
- From the Centre for Protein Science and Crystallography, School of Life Sciences.
Organizational Affiliation: