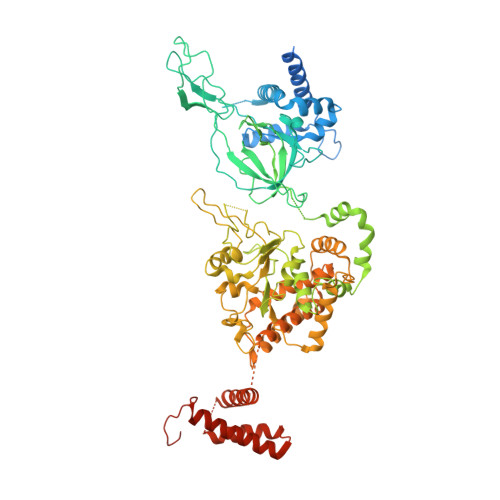
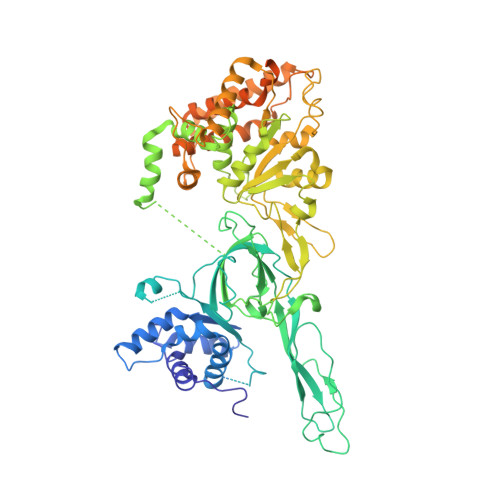
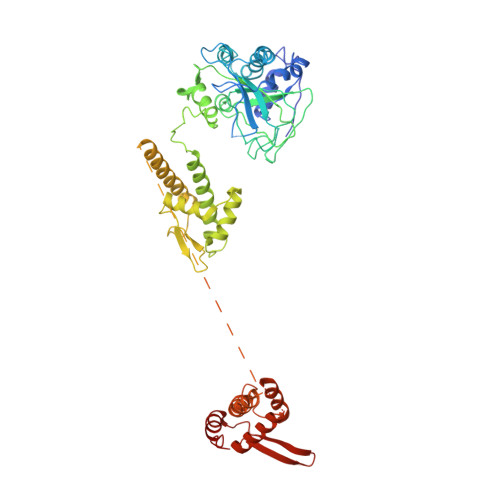
Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer
Zhai, Y., Cheng, E., Wu, H., Li, N., Yung, P.Y., Gao, N., Tye, B.K.(2017) Nat Struct Mol Biol 24: 300-308
- PubMed: 28191894
- DOI: https://doi.org/10.1038/nsmb.3374
- Primary Citation of Related Structures:
5XF8 - PubMed Abstract:
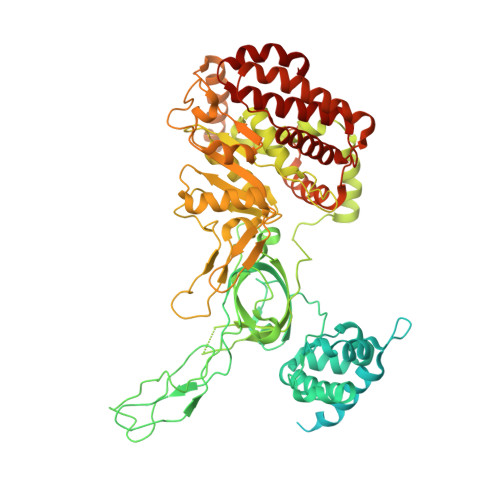
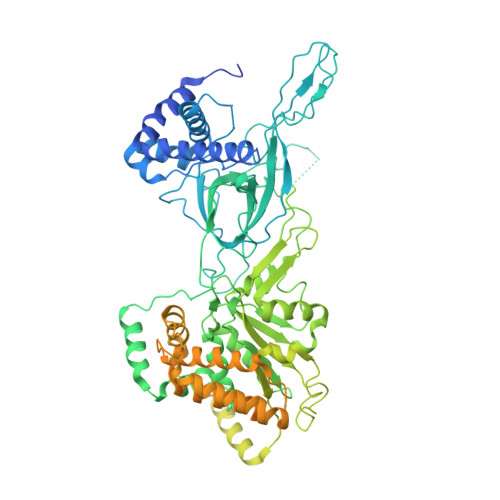
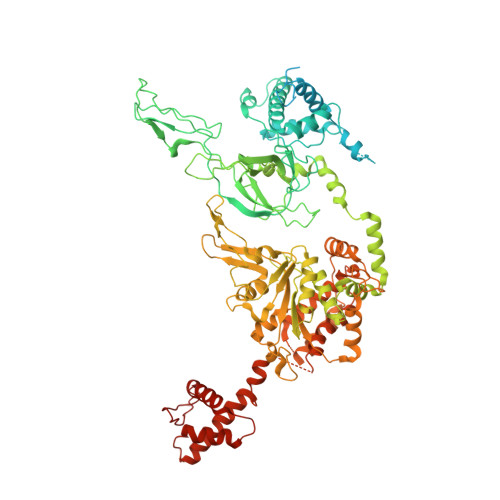
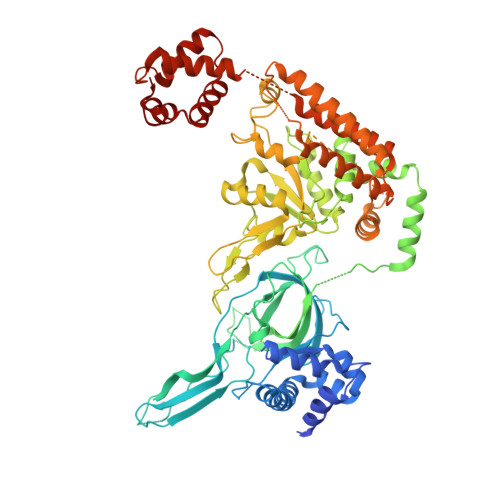
The minichromosome maintenance complex (MCM) hexameric complex (Mcm2-7) forms the core of the eukaryotic replicative helicase. During G1 phase, two Cdt1-Mcm2-7 heptamers are loaded onto each replication origin by the origin-recognition complex (ORC) and Cdc6 to form an inactive MCM double hexamer (DH), but the detailed loading mechanism remains unclear. Here we examine the structures of the yeast MCM hexamer and Cdt1-MCM heptamer from Saccharomyces cerevisiae. Both complexes form left-handed coil structures with a 10-15-Å gap between Mcm5 and Mcm2, and a central channel that is occluded by the C-terminal domain winged-helix motif of Mcm5. Cdt1 wraps around the N-terminal regions of Mcm2, Mcm6 and Mcm4 to stabilize the whole complex. The intrinsic coiled structures of the precursors provide insights into the DH formation, and suggest a spring-action model for the MCM during the initial origin melting and the subsequent DNA unwinding.
- Division of Life Science, Hong Kong University of Science and Technology, Clear Water Bay, Hong Kong, China.
Organizational Affiliation: