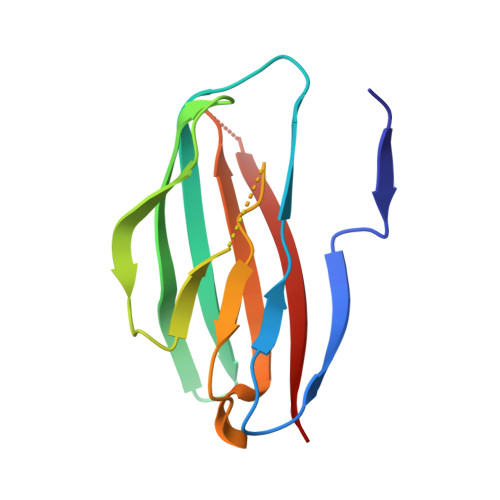
Parallel homodimer structures of the extracellular domains of the voltage-gated sodium channel beta 4 subunit explain its role in cell-cell adhesion
Shimizu, H., Tosaki, A., Ohsawa, N., Ishizuka-Katsura, Y., Shoji, S., Miyazaki, H., Oyama, F., Terada, T., Shirouzu, M., Sekine, S.I., Nukina, N., Yokoyama, S.(2017) J Biological Chem 292: 13428-13440
- PubMed: 28655765
- DOI: https://doi.org/10.1074/jbc.M117.786509
- Primary Citation of Related Structures:
5XAW, 5XAX - PubMed Abstract:
Voltage-gated sodium channels (VGSCs) are transmembrane proteins required for the generation of action potentials in excitable cells and essential for propagating electrical impulses along nerve cells. VGSCs are complexes of a pore-forming α subunit and auxiliary β subunits, designated as β1/β1B-β4 (encoded by SCN1B-4B, respectively), which also function in cell-cell adhesion. We previously reported the structural basis for the trans homophilic interaction of the β4 subunit, which contributes to its adhesive function. Here, using crystallographic and biochemical analyses, we show that the β4 extracellular domains directly interact with each other in a parallel manner that involves an intermolecular disulfide bond between the unpaired Cys residues (Cys 58 ) in the loop connecting strands B and C and intermolecular hydrophobic and hydrogen-bonding interactions of the N-terminal segments (Ser 30 -Val 35 ). Under reducing conditions, an N-terminally deleted β4 mutant exhibited decreased cell adhesion compared with the wild type, indicating that the β4 cis dimer contributes to the trans homophilic interaction of β4 in cell-cell adhesion. Furthermore, this mutant exhibited increased association with the α subunit, indicating that the cis dimerization of β4 affects α-β4 complex formation. These observations provide the structural basis for the parallel dimer formation of β4 in VGSCs and reveal its mechanism in cell-cell adhesion.
- From the RIKEN Systems and Structural Biology Center, Tsurumi, Yokohama 230-0045, Japan.
Organizational Affiliation: