Structural mimicry of the dengue virus envelope glycoprotein revealed by the crystallographic study of an idiotype-anti-idiotype Fab complex
Wong, Y.H., Goh, B.C., Lim, S.Y., Teo, E.W., Lim, A.P.C., Dedon, P.C., Hanson, B.J., MacAry, P.A., Lescar, J.(2017) J Virol
- PubMed: 28637753
- DOI: https://doi.org/10.1128/JVI.00406-17
- Primary Citation of Related Structures:
5XAJ - PubMed Abstract:
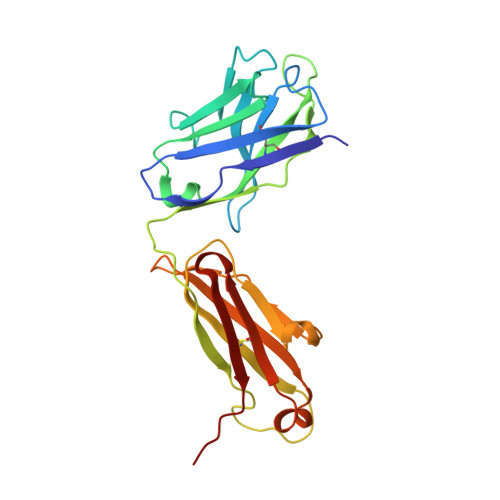
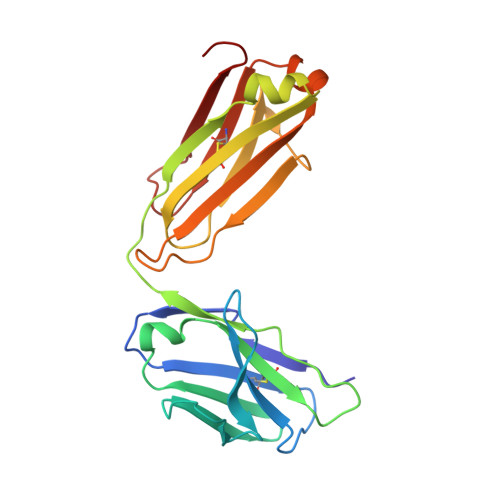
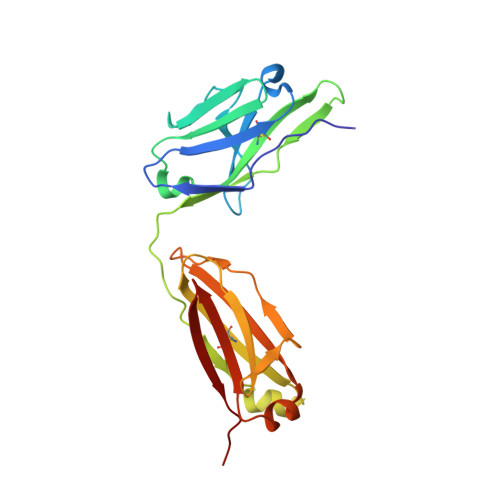
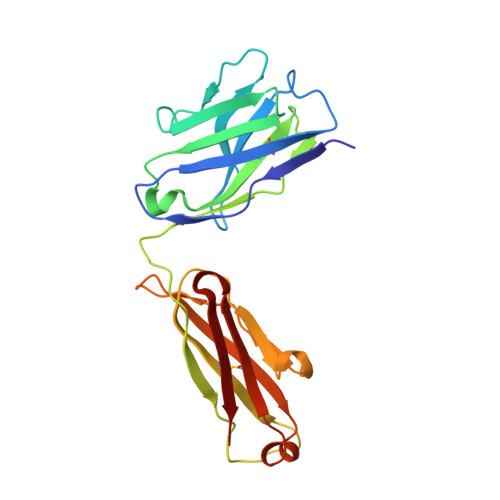
A detailed understanding of the fine specificity of serotype-specific human antibodies is vital for the development and evaluation of new vaccines for pathogenic flaviviruses such as dengue virus (DENV) and Zika virus. In this study, we thoroughly characterize the structural footprint of an anti-idiotype antibody (E1) specific for a potent, fully human DENV serotype 1-specific antibody, termed HM14c10, derived from a recovered patient. The crystal structure at a resolution of 2.5 Å of a complex between the Fab fragments of E1 and HM14c10 provides the first detailed molecular comparison of an anti-idiotype paratope specific for a human antibody with its analogous epitope, a discontinuous quaternary structure located at the surface of the viral particle that spans adjacent envelope (E) proteins. This comparison reveals that the footprints left by E1 and E on HM14c10 largely overlap, explaining why the formation of binary complexes is mutually exclusive. Structural mimicry of the DENV E epitope by the E1 combining site is achieved via the formation of numerous interactions with heavy chain complementarity domain regions (CDRs) of HM14c10, while fewer interactions are observed with its light chain than for the E protein. We show that E1 can be utilized to detect HM14c10-like antibodies in sera from patients who recovered from DENV-1, infection suggesting that this is a public (common) idiotype. These data demonstrate the utility of employing an anti-idiotype antibody to monitor a patient's specific immune responses and suggest routes for the improvement of E "mimicry" by E1 by increasing its recognition of the Fab HM14c10 light chain CDRs. IMPORTANCE A chimeric yellow fever-dengue live-attenuated tetravalent vaccine is now being marketed. Dengue remains a significant public health problem, because protection conferred by this vaccine against the four circulating serotypes is uneven. Reliable tools must be developed to measure the immune responses of individuals exposed to DENV either via viral infection or through vaccination. Anti-idiotypic antibodies provide precision tools for analyzing the pharmacokinetics of antibodies in an immune response and also for measuring the amount of circulating anti-infective therapeutic antibodies. Here, we characterize how an anti-idiotypic antibody (E1) binds antibody HM14c10, which potently neutralizes DENV serotype 1. We report the crystal structure at a resolution of 2.5 Å of a complex between the Fab fragments of E1 and HM14c10 and provide the first detailed molecular comparison between the anti-idiotype surface and its analogous epitope located at the surface of the dengue virus particle.
- Nanyang Institute for Structural Biology, School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.
Organizational Affiliation: