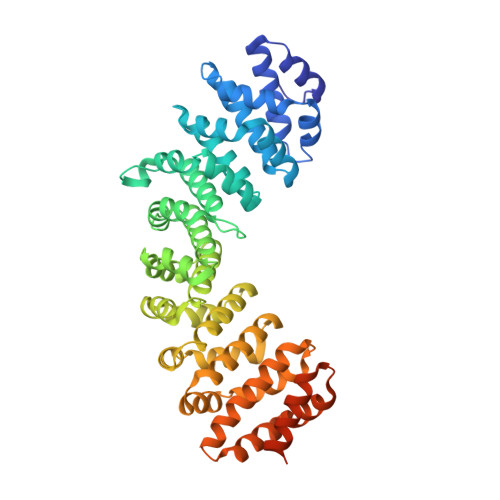
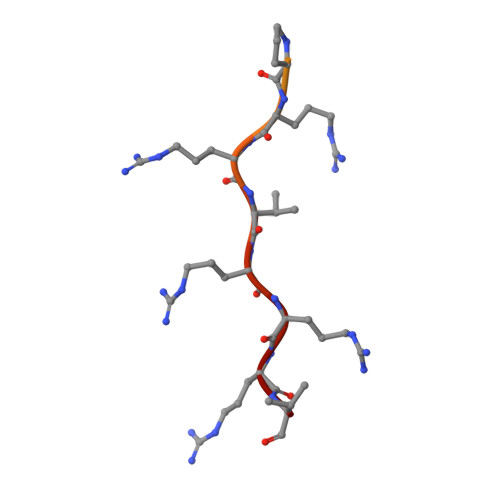
Crystal structure of importin-alpha bound to the nuclear localization signal of Epstein-Barr virus EBNA-LP protein
Nakada, R., Matsuura, Y.(2017) Protein Sci 26: 1231-1235
- PubMed: 28383161
- DOI: https://doi.org/10.1002/pro.3173
- Primary Citation of Related Structures:
5X8N - PubMed Abstract:
Epstein-Barr virus EBNA-LP protein is a transcriptional coactivator of EBNA2. Efficient nuclear localization of EBNA-LP is essential for cooperation with EBNA2. Here, we report the crystal structure of the nuclear import adaptor importin-α1 bound to the nuclear localization signal (NLS) of EBNA-LP that shows EBNA-LP residues 44-RRVRRR-49 binding to the major NLS-binding site at the P0-P5 positions. In contrast to previously characterized classical NLSs that invariably have a basic residue [either lysine (in the vast majority of cases) or arginine] at the P2 position, the EBNA-LP NLS is unique in that it has valine at the P2 position. The loss of the critical P2 lysine (or arginine) is compensated by arginine at the P0 position in the EBNA-LP NLS.
- Division of Biological Science, Graduate School of Science, Nagoya University, Japan.
Organizational Affiliation: