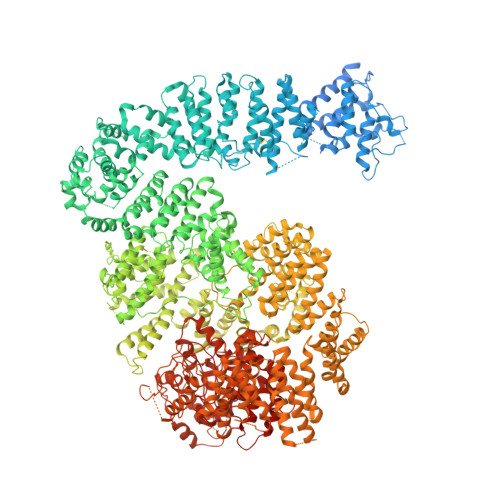
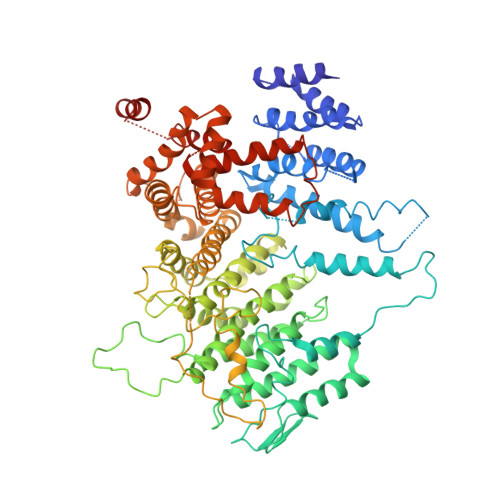
3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Wang, X., Ran, T., Zhang, X., Xin, J., Zhang, Z., Wu, T., Wang, W., Cai, G.(2017) Science 358: 1206-1209
- PubMed: 29191911
- DOI: https://doi.org/10.1126/science.aan8414
- Primary Citation of Related Structures:
5X6O - PubMed Abstract:
The ataxia telangiectasia-mutated and Rad3-related (ATR) kinase is a master regulator of DNA damage response and replication stress in humans, but the mechanism of its activation remains unclear. ATR acts together with its partner ATRIP. Using cryo-electron microscopy, we determined the structure of intact Mec1-Ddc2 (the yeast homolog of ATR-ATRIP), which is poised for catalysis, at a resolution of 3.9 angstroms. Mec1-Ddc2 forms a dimer of heterodimers through the PRD and FAT domains of Mec1 and the coiled-coil domain of Ddc2. The PRD and Bridge domains in Mec1 constitute critical regulatory sites. The activation loop of Mec1 is inhibited by the PRD, revealing an allosteric mechanism of kinase activation. Our study clarifies the architecture of ATR-ATRIP and provides a structural framework for the understanding of ATR regulation.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei 230027, China.
Organizational Affiliation: