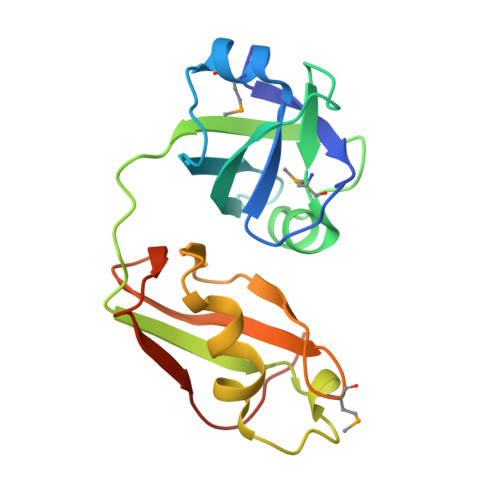
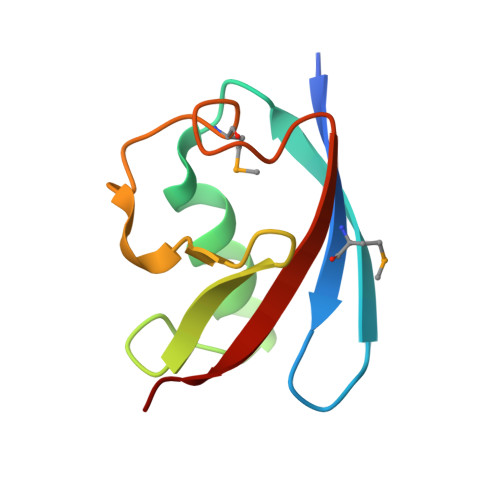
Crystal structures of the UBX domain of human UBXD7 and its complex with p97 ATPase
Li, Z.H., Wang, Y., Xu, M., Jiang, T.(2017) Biochem Biophys Res Commun 486: 94-100
- PubMed: 28274878
- DOI: https://doi.org/10.1016/j.bbrc.2017.03.005
- Primary Citation of Related Structures:
5X3P, 5X4L - PubMed Abstract:
In humans, UBXD7 (also called UBXN7), an adaptor of p97 ATPase, can participate in the degradation of misfolded or damaged proteins in the p97-mediated ubiquitin proteasome system (UPS). UBXD7 binds to ubiquitinated substrates via its UBA domain and interacts with p97 N-terminal domain through its UBX domain to recruit p97 or the p97 core complex (p97/NPL4/UFD1). Here, we report the crystal structures of the UBX domain of UBXD7 (UBXD7 UBX ) at 2.0 Å resolution and its complex with p97 N-terminal domain (p97 NTD -UBXD7 UBX complex) at 2.4 Å resolution. A structural analysis and isothermal titration calorimetry results provide detailed molecular basis of interaction between UBXD7 UBX and p97 NTD . Moreover, structural superpositions suggest that dimerization of UBXD7 UBX via an intermolecular disulfide bond could interfere with the formation of the p97 NTD -UBXD7 UBX complex. Interestingly, UBXD7 may have a cooperative effect on p97 interaction with UFD1. Together, these results provide structural and biochemical insights into the interaction between p97 NTD and UBXD7 UBX .
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, 19A Yuquan Road, Shijingshan District, Beijing 100049, China.
Organizational Affiliation: