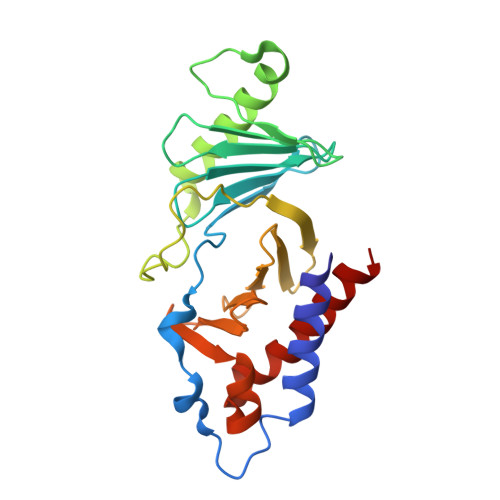
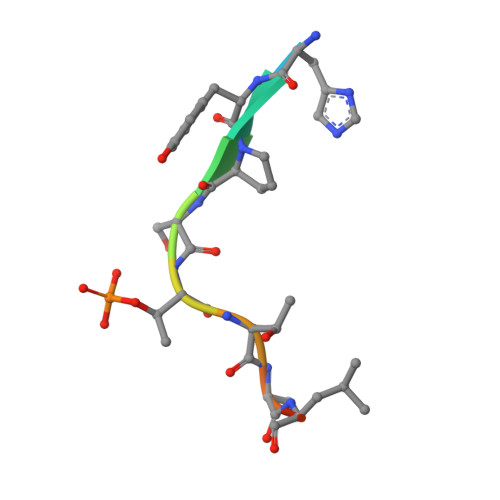
Phosphorylation of human enhancer filamentation 1 (HEF1) stimulates interaction with Polo-like kinase 1 leading to HEF1 localization to focal adhesions.
Lee, K.H., Hwang, J.A., Kim, S.O., Kim, J.H., Shin, S.C., Kim, E.E., Lee, K.S., Rhee, K., Jeon, B.H., Bang, J.K., Cha-Molstad, H., Soung, N.K., Jang, J.H., Ko, S.K., Lee, H.G., Ahn, J.S., Kwon, Y.T., Kim, B.Y.(2018) J Biological Chem 293: 847-862
- PubMed: 29191835
- DOI: https://doi.org/10.1074/jbc.M117.802587
- Primary Citation of Related Structures:
5X3S - PubMed Abstract:
Elevated expression of human enhancer filamentation 1 (HEF1; also known as NEDD9 or Cas-L) is an essential stimulus for the metastatic process of various solid tumors. This process requires HEF1 localization to focal adhesions (FAs). Although the association of HEF1 with FAs is considered to play a role in cancer cell migration, the mechanism targeting HEF1 to FAs remains unclear. Moreover, up-regulation of Polo-like kinase 1 (Plk1) positively correlates with human cancer metastasis, yet how Plk1 deregulation promotes metastasis remains elusive. Here, we report that casein kinase 1δ (CK1δ) phosphorylates HEF1 at Ser-780 and Thr-804 and that these phosphorylation events promote a physical interaction between Plk1 and HEF1. We found that this interaction is critical for HEF1 translocation to FAs and for inducing migration of HeLa cells. Plk1-docking phosphoepitopes were mapped/confirmed in HEF1 by various methods, including X-ray crystallography, and mutated for functional analysis in HeLa cells. In summary, our results reveal the role of a phosphorylation-dependent HEF1-Plk1 complex in HEF1 translocation to FAs to induce cell migration. Our findings provide critical mechanistic insights into the HEF1-Plk1 complex-dependent localization of HEF1 to FAs underlying the metastatic process and may therefore contribute to the development of new cancer therapies.
- From the World Class Institute, Anticancer Agent Research Center, Korea Research Institute of Bioscience and Biotechnology, 30 Yeongudanji-ro, Ochang, Cheongwon, Chungbuk 28116, Korea, leekh@kribb.re.kr.
Organizational Affiliation: