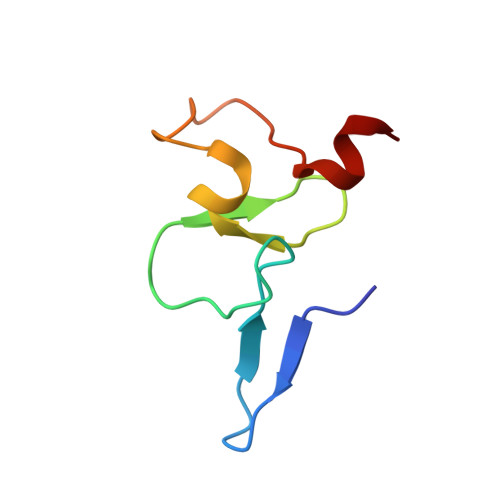
Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Zhao, S., Yang, M., Zhou, W., Zhang, B., Cheng, Z., Huang, J., Zhang, M., Wang, Z., Wang, R., Chen, Z., Zhu, J., Li, H.(2017) Proc Natl Acad Sci U S A 114: E7245-E7254
- PubMed: 28808021
- DOI: https://doi.org/10.1073/pnas.1704155114
- Primary Citation of Related Structures:
5WXG, 5WXH - PubMed Abstract:
Chemical modifications on histones and DNA/RNA constitute a fundamental mechanism for epigenetic regulation. These modifications often function as docking marks to recruit or stabilize cognate "reader" proteins. So far, a platform for quantitative and high-throughput profiling of the epigenetic interactome is urgently needed but still lacking. Here, we report a 3D-carbene chip-based surface plasmon resonance imaging (SPRi) technology for this purpose. The 3D-carbene chip is suitable for immobilizing versatile biomolecules (e.g., peptides, antibody, DNA/RNA) and features low nonspecific binding, random yet function-retaining immobilization, and robustness for reuses. We systematically profiled binding kinetics of 1,000 histone "reader-mark" pairs on a single 3D-carbene chip and validated two recognition events by calorimetric and structural studies. Notably, a discovery on H3K4me3 recognition by the DNA mismatch repair protein MSH6 in Capsella rubella suggests a mechanism of H3K4me3-mediated DNA damage repair in plant.
- Ministry of Education Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Organizational Affiliation: