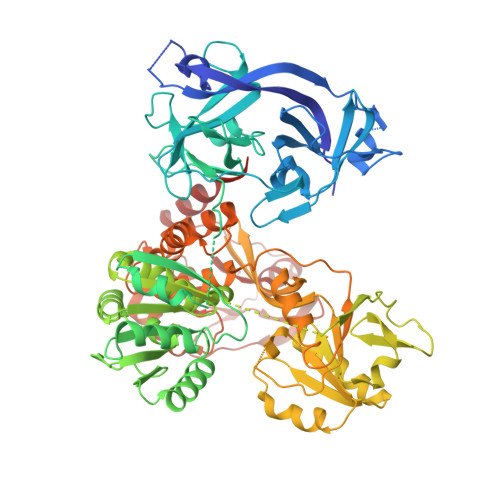
Uncoupling of Protease trans-Cleavage and Helicase Activities in Pestivirus NS3.
Zheng, F., Lu, G., Li, L., Gong, P., Pan, Z.(2017) J Virol 91
- PubMed: 28835495
- DOI: https://doi.org/10.1128/JVI.01094-17
- Primary Citation of Related Structures:
5WX1 - PubMed Abstract:
The nonstructural protein NS3 from the Flaviviridae family is a multifunctional protein that contains an N-terminal protease and a C-terminal helicase, playing essential roles in viral polyprotein processing and genome replication. Here we report a full-length crystal structure of the classical swine fever virus (CSFV) NS3 in complex with its NS4A protease cofactor segment (PCS) at a 2.35-Å resolution. The structure reveals a previously unidentified ∼2,200-Å 2 intramolecular protease-helicase interface comprising three clusters of interactions, representing a "closed" global conformation related to the NS3-NS4A cis -cleavage event. Although this conformation is incompatible with protease trans -cleavage, it appears to be functionally important and beneficial to the helicase activity, as the mutations designed to perturb this conformation impaired both the helicase activities in vitro and virus production in vivo Our work reveals important features of protease-helicase coordination in pestivirus NS3 and provides a key basis for how different conformational states may explicitly contribute to certain functions of this natural protease-helicase fusion protein. IMPORTANCE Many RNA viruses encode helicases to aid their RNA genome replication and transcription by unwinding structured RNA. Being naturally fused to a protease participating in viral polyprotein processing, the NS3 helicases encoded by the Flaviviridae family viruses are unique. Therefore, how these two enzyme modules coordinate in a single polypeptide is of particular interest. Here we report a previously unidentified conformation of pestivirus NS3 in complex with its NS4A protease cofactor segment (PCS). This conformational state is related to the protease cis -cleavage event and is optimal for the function of helicase. This work provides an important basis to understand how different enzymatic activities of NS3 may be achieved by the coordination between the protease and helicase through different conformational states.
- State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan, Hubei, China.
Organizational Affiliation: