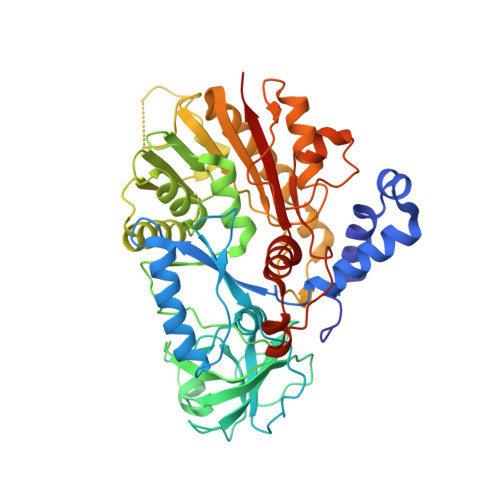
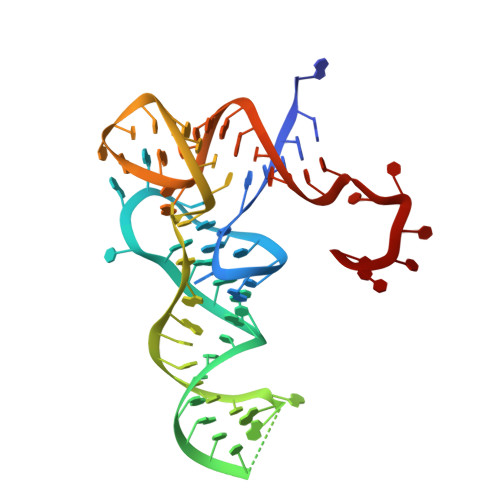
Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6
Liu, R.J., Long, T., Li, J., Li, H., Wang, E.D.(2017) Nucleic Acids Res 45: 6684-6697
- PubMed: 28531330
- DOI: https://doi.org/10.1093/nar/gkx473
- Primary Citation of Related Structures:
5WWQ, 5WWR, 5WWS, 5WWT - PubMed Abstract:
5-methylcytosine (m5C) modifications of RNA are ubiquitous in nature and play important roles in many biological processes such as protein translational regulation, RNA processing and stress response. Aberrant expressions of RNA:m5C methyltransferases are closely associated with various human diseases including cancers. However, no structural information for RNA-bound RNA:m5C methyltransferase was available until now, hindering elucidation of the catalytic mechanism behind RNA:m5C methylation. Here, we have solved the structures of NSun6, a human tRNA:m5C methyltransferase, in the apo form and in complex with a full-length tRNA substrate. These structures show a non-canonical conformation of the bound tRNA, rendering the base moiety of the target cytosine accessible to the enzyme for methylation. Further biochemical assays reveal the critical, but distinct, roles of two conserved cysteine residues for the RNA:m5C methylation. Collectively, for the first time, we have solved the complex structure of a RNA:m5C methyltransferase and addressed the catalytic mechanism of the RNA:m5C methyltransferase family, which may allow for structure-based drug design toward RNA:m5C methyltransferase-related diseases.
- State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, P. R. China.
Organizational Affiliation: